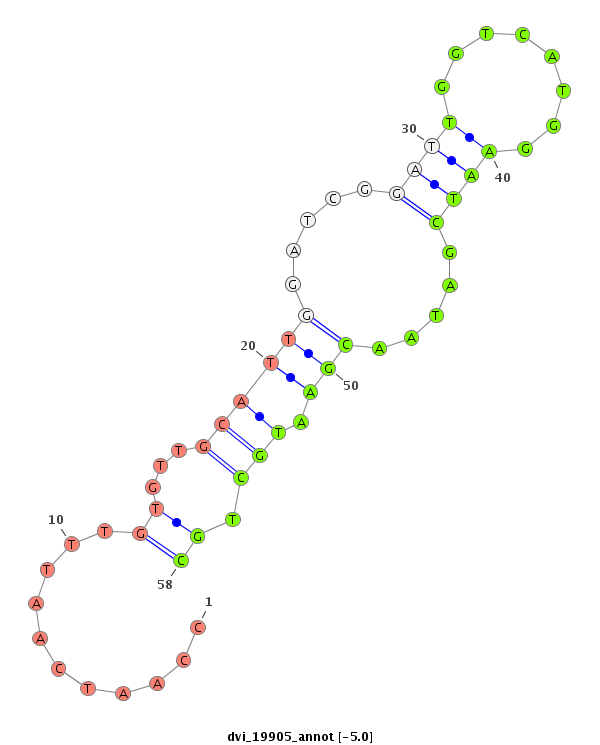
ID:dvi_19905 |
Coordinate:scaffold_13049:13948688-13948838 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
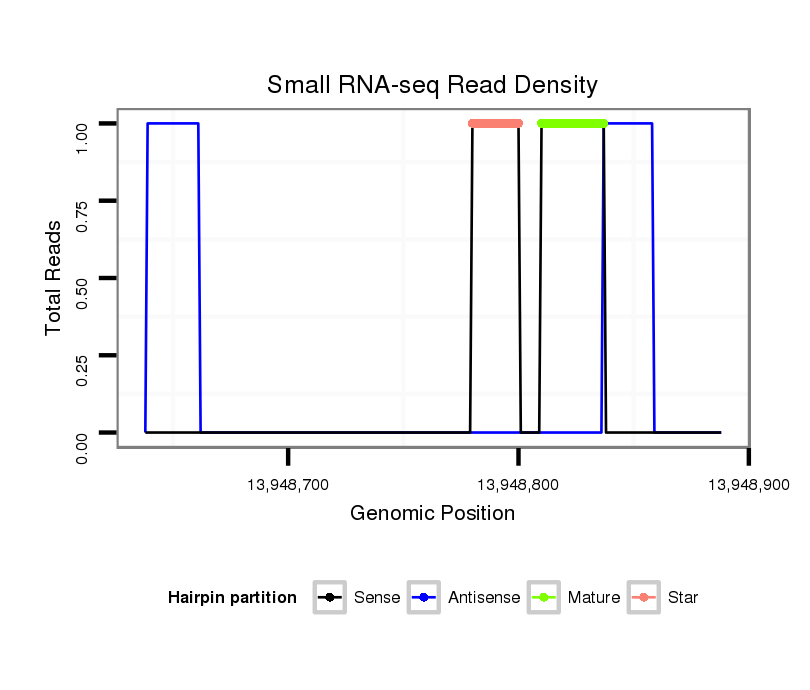
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -12.3 |
 |
CDS [Dvir\GJ13553-cds]; exon [dvir_GLEANR_13436:1]; intron [Dvir\GJ13553-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CGTTTTGGAGCTTGTCGTGCAGAAAACTTAAAAATGTAATTATTTACAATGTAAGTGTTCCTTTTATGAACTTCTATAGACTAGCCACGATATGCTAAGTTATCAACAATCATATTTACTCAGTGACCGTTCAGATGGATGGCCAATCAATTTGTGTTGCATTGGATCGGATTGGTCATGGAATCGATAACGAATGCTGCGTCGTCAGTCGGCAGTCCAAATAAACCGAAATGAGTAGAGCATCTCTCAAA **********************************************************************************************************************************************...........((...((((((.....((((........)))).....))).))).))*************************************************** |
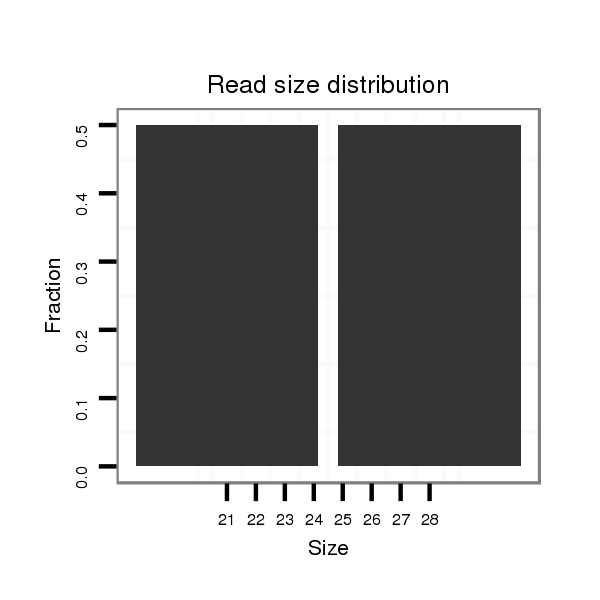
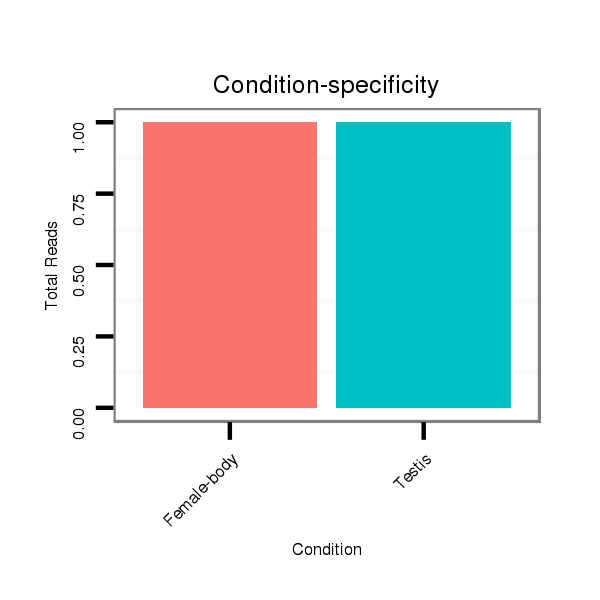
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
SRR060681 Argx9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
M027 male body |
M061 embryo |
SRR060687 9_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................GACTAACGACGATATTCTA.......................................................................................................................................................... | 19 | 3 | 13 | 2.00 | 26 | 0 | 0 | 0 | 10 | 8 | 6 | 0 | 0 | 1 | 0 | 1 | 0 |
| ............................................................................................................................................................................TGGTCATGGAATCGATAACGAATGCTGC................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CCAATCAATTTGTGTTGCATT........................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TTCGAGAACGAATGCTGCGTGGT.............................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ATCGAGAACGAATTCTGCGTGGT.............................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ATCGGAAACGAATGCTGCGTGGT.............................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AGAACGAATGCTGCGTGGT.............................................. | 19 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GAATTAGATCGGATTGGTC.......................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CCGGGTTGGTCATGGGATC.................................................................. | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TTCGAGAACGAATTCTGCGT................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................ATAACGAATCCTGGGTGGT.............................................. | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GGTTCGGATTGGTTATGG...................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................CGGATCGGATTGGTCTCG....................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................CGATAACGAATACGGCTTC................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
GCAAAACCTCGAACAGCACGTCTTTTGAATTTTTACATTAATAAATGTTACATTCACAAGGAAAATACTTGAAGATATCTGATCGGTGCTATACGATTCAATAGTTGTTAGTATAAATGAGTCACTGGCAAGTCTACCTACCGGTTAGTTAAACACAACGTAACCTAGCCTAACCAGTACCTTAGCTATTGCTTACGACGCAGCAGTCAGCCGTCAGGTTTATTTGGCTTTACTCATCTCGTAGAGAGTTT
***************************************************...........((...((((((.....((((........)))).....))).))).))********************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
M027 male body |
V047 embryo |
SRR060671 9x160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................GTGACTGGCAAGTCTACCTAC.............................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .CAAAACCTCGAACAGCACGTCTT................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................GCAGCAGTCAGCCGTCAGGTTT.............................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................CTTAGCTCTGGCTTACGA..................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................GTCCCTTAGCTATCGCTT......................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................CTGCTGGTCAGTTAAACAC............................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................TGGCAAGTCAACCTGCTG............................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:13948638-13948888 + | dvi_19905 | CGTTTTGGAGCTTGTCGTGCAGAAAACTTAAAAATGTAATTATTTACAATGTAAGTGTTCCTTTTATGAACTTCTATAGACTAGCCACGATATGCTAAGTTATCAACAATCATATTTACTCAGTGACCGTTCAGATGGATGGCCAATCAATTTGTGTTGCATTGGATCGGATTGGTCATGGAATCGATAACGAATGCTGCGTCGTCAGTCGGCAGTCCAAATAAACCGAAATGAGTAGAGCATCTCTCAAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 04:33 AM