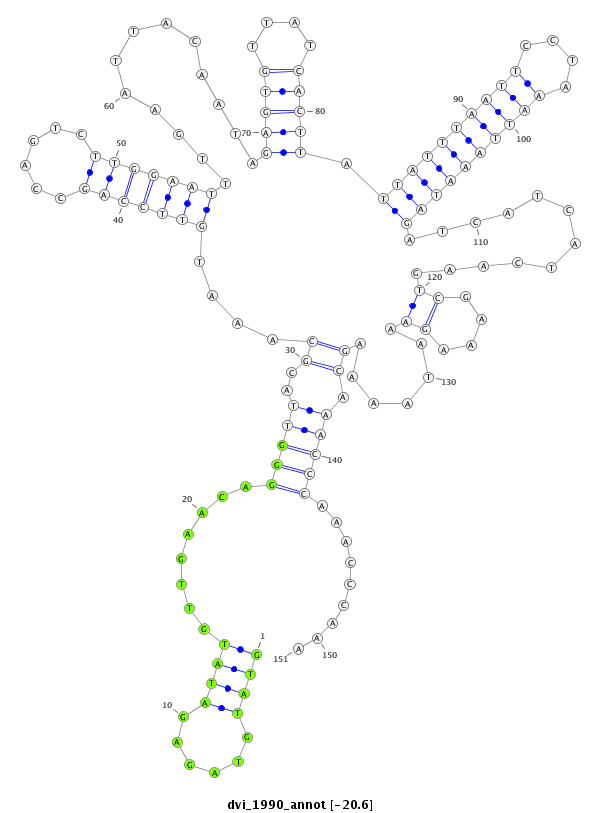
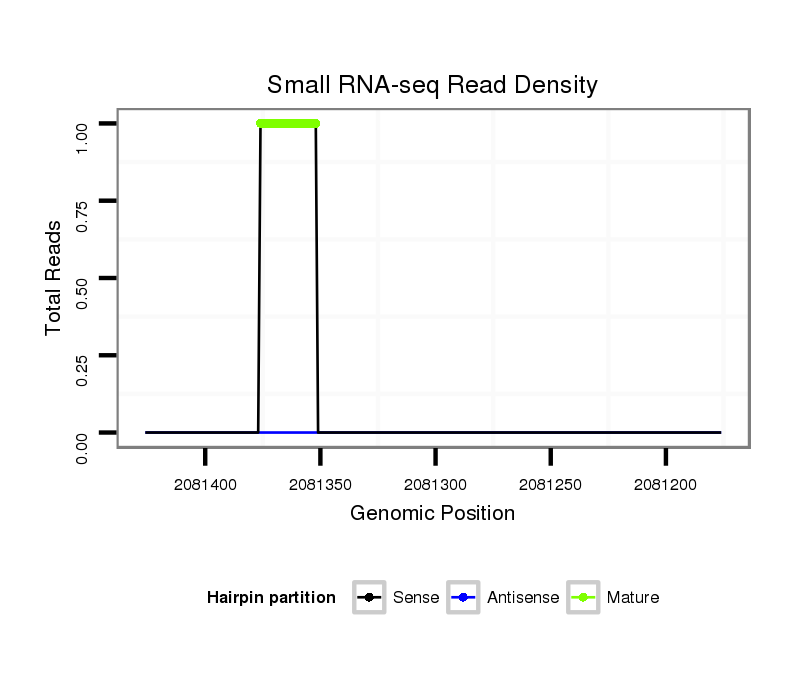
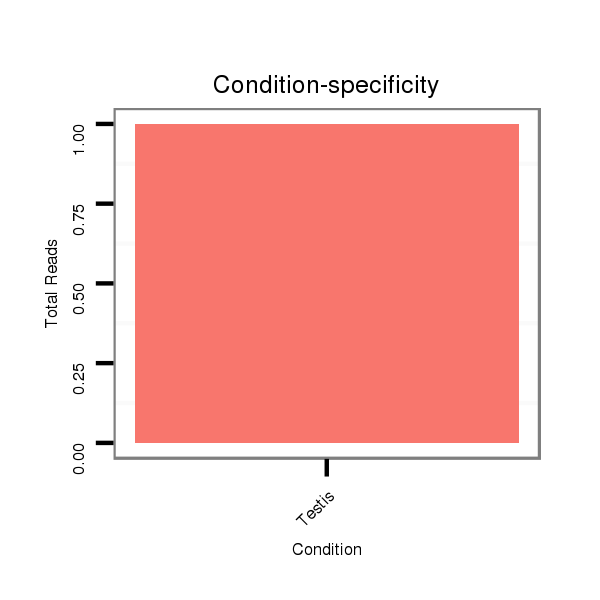
ID:dvi_1990 |
Coordinate:scaffold_12726:2081226-2081376 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -20.4 | -20.2 | -20.1 |
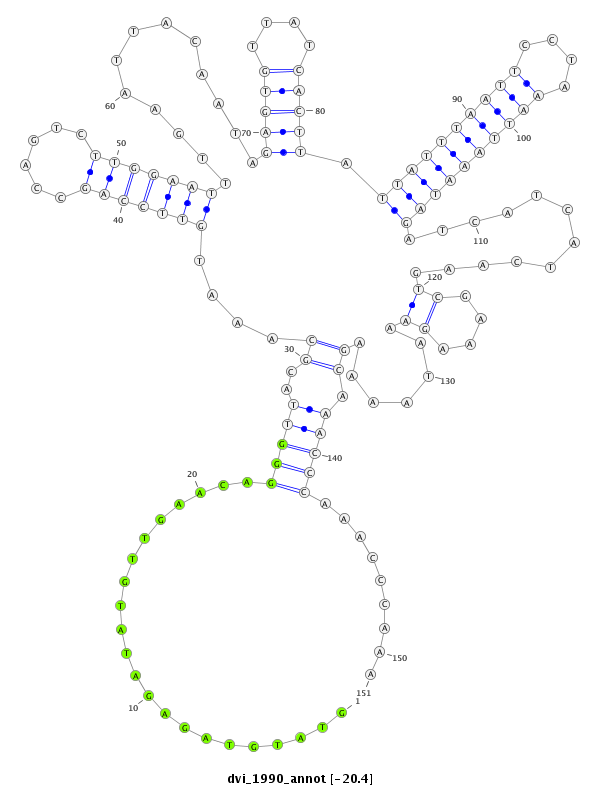 |
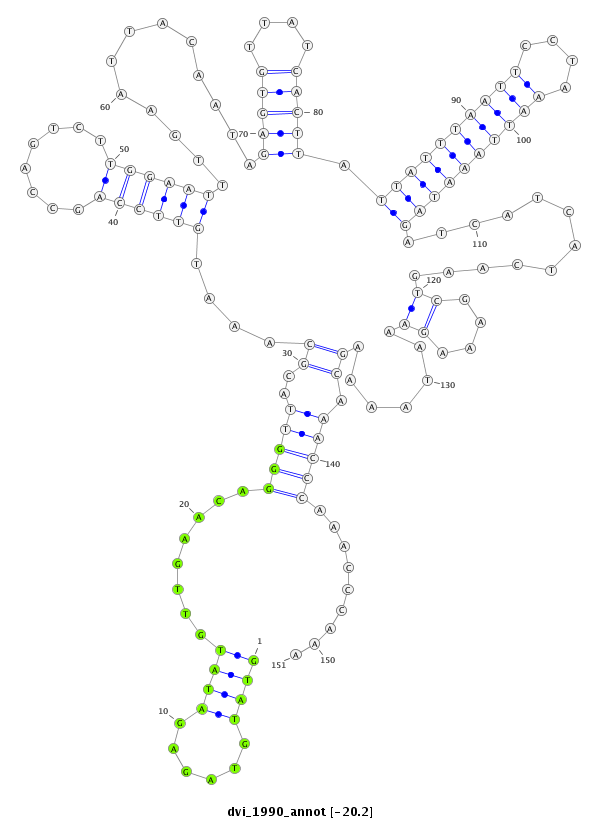 |
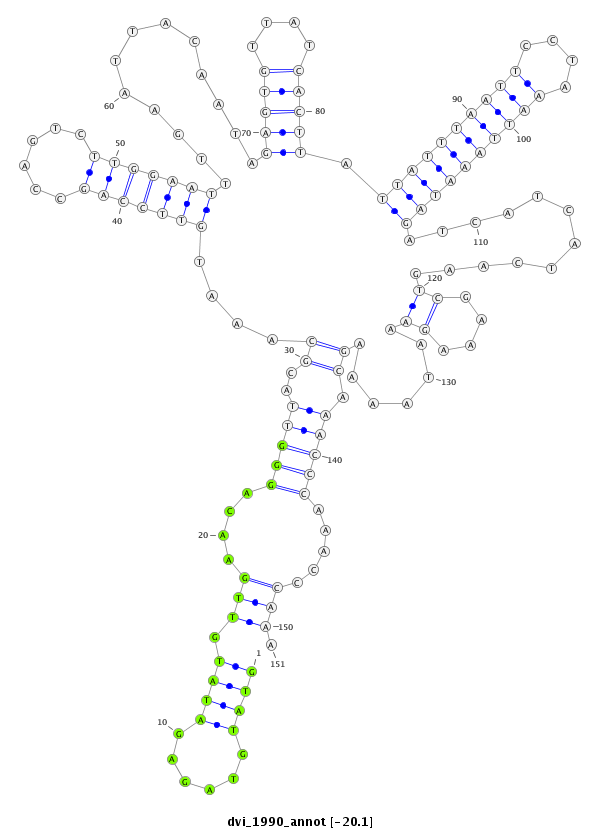 |
CDS [Dvir\GJ15200-cds]; exon [dvir_GLEANR_15525:7]; intron [Dvir\GJ15200-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTGAAATTAATACGCCCGATATCGCAGATATTAAATTAACATCGCCCAAGGTATGTAGAGATATGTTGAACAGGGTTACGCAAATGTTCCAGCCAGTCTTGGAATTTGAATTACAATAGAGTGTTATCACTTATTATTTAATTCCTAAATTAAATAGATCATCATCAAGTCGAAAGAAATAAAAGCAAACCCAAACCCAAAGGCCGAACGTAGCCTATTTACCAAGGCAAAGGATGAAAAACCAGTTAGTA **************************************************((((......))))........(((((..((....(((((((......))))))).............(((((....))))).((((((((((....))))))))))............((....)).......)).))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060658 140_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060659 Argentina_testes_total |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGTAGAGATATGTTGAACAGGG................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................CCAGTCTTGTAATTGGAATGA.......................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................ATGTTACCATCGCCCAAGGT....................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................CAGTCTTGTAATTGGAATGA.......................................................................................................................................... | 20 | 3 | 18 | 0.11 | 2 | 0 | 1 | 0 | 1 | 0 |
| .......................................CATCGCCCAAAATATGTACA................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
AACTTTAATTATGCGGGCTATAGCGTCTATAATTTAATTGTAGCGGGTTCCATACATCTCTATACAACTTGTCCCAATGCGTTTACAAGGTCGGTCAGAACCTTAAACTTAATGTTATCTCACAATAGTGAATAATAAATTAAGGATTTAATTTATCTAGTAGTAGTTCAGCTTTCTTTATTTTCGTTTGGGTTTGGGTTTCCGGCTTGCATCGGATAAATGGTTCCGTTTCCTACTTTTTGGTCAATCAT
**************************************************((((......))))........(((((..((....(((((((......))))))).............(((((....))))).((((((((((....))))))))))............((....)).......)).))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060664 9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................CTTTCTTAATTTACGTTGGGG........................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ................................................TCTATATAACTCTATACAACTT..................................................................................................................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................AGTAGTAGTGCAACTATCTT......................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................TCCGATGAATGGCTCCGTT..................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:2081176-2081426 - | dvi_1990 | TTGAAATTAATACGCCCGATATCGCAGATATTAAATTAACATCGCCCAAGGTATGTAGAGATATGTTGAACAGGGTTACGCAAATGTTCCAGCCAGTCTTGGAATTTGAATTACAATAGAGTGTTATCACTTATTATTTAATTCCTAAATTAAATAGATCATCATCAAGTCGAAAGAAATAAAAGCAAACCCAAACCCAAAGGCCGAACGTAGCCTATTTACCAAGGCAAAGGATGAAAAACCAGTTAGTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 04:41 AM