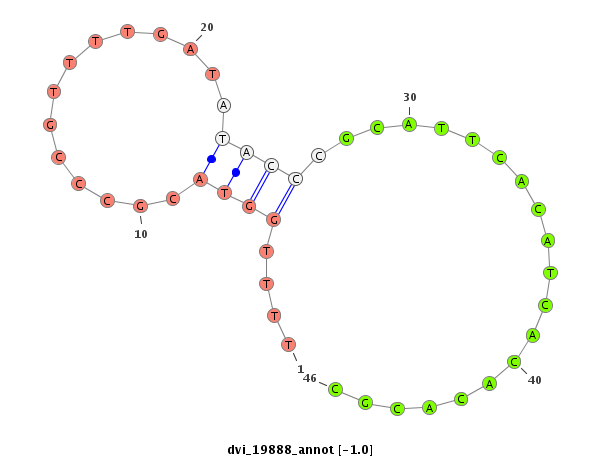
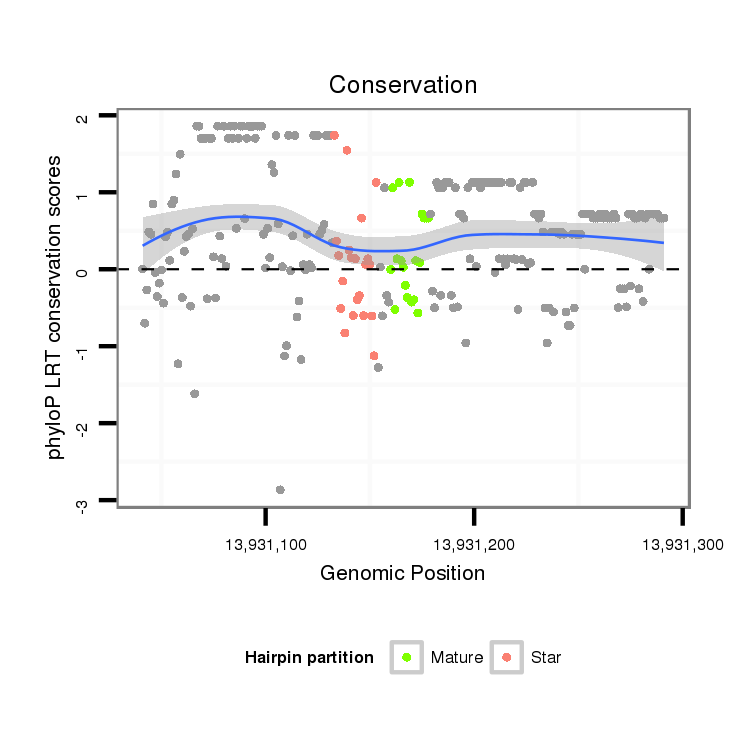
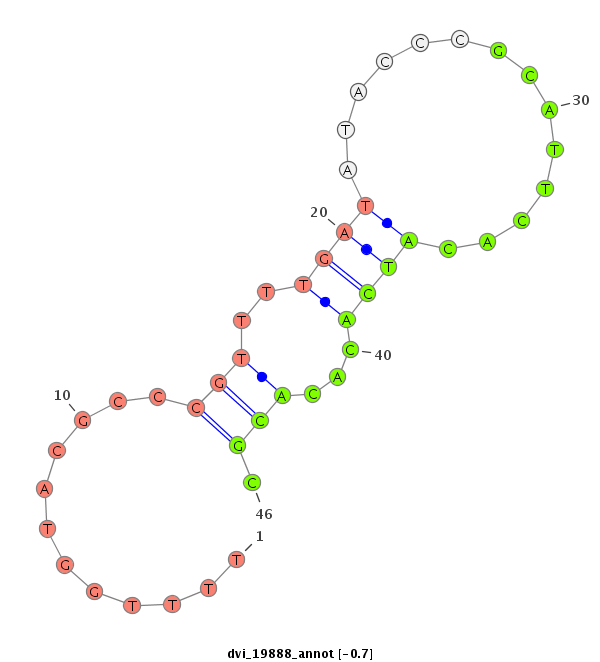
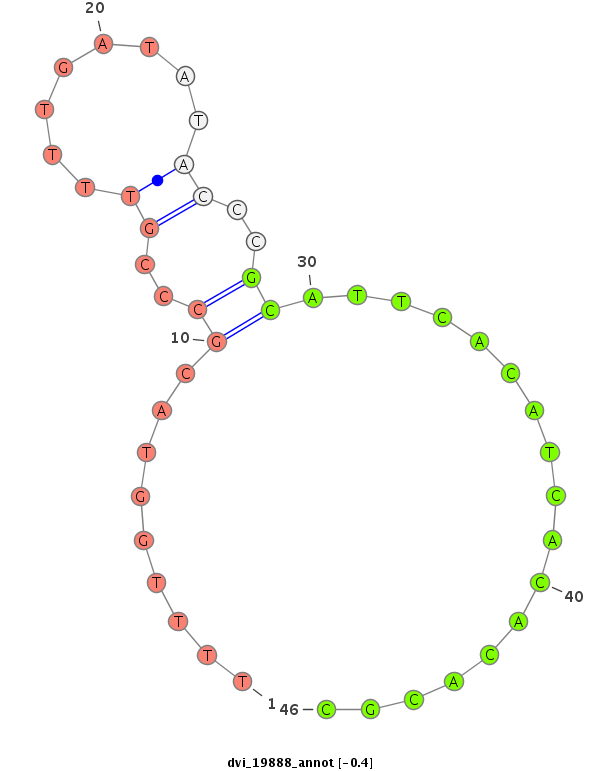
ID:dvi_19888 |
Coordinate:scaffold_13049:13931091-13931241 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -0.7 | -0.5 | -0.4 |
 |
 |
 |
CDS [Dvir\GJ13551-cds]; exon [dvir_GLEANR_13434:1]; intron [Dvir\GJ13551-in]
No Repeatable elements found
| --------------------------########################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTTAGCAGCGACTTCAGCCCTTGGAAATGGCGCGTTTAATTGAGAGCAATGTAAGTTACGAAACAAACTGTGCAGCTCAGCTTTTGCATAATTTTTGGTACGCCCGTTTTGATATACCCGCATTCACATCACACACGCATATCACATTTGGCTAGGCTATTTCATAATTCTTAAAAATATACATTTGTATGTACGCATATGTGTGGCTGCCATTTTGGCATCTCCCACAAAATAATGCAATGAAATATGCG ********************************************************************************************....((((..............))))....................***************************************************************************************************************** |
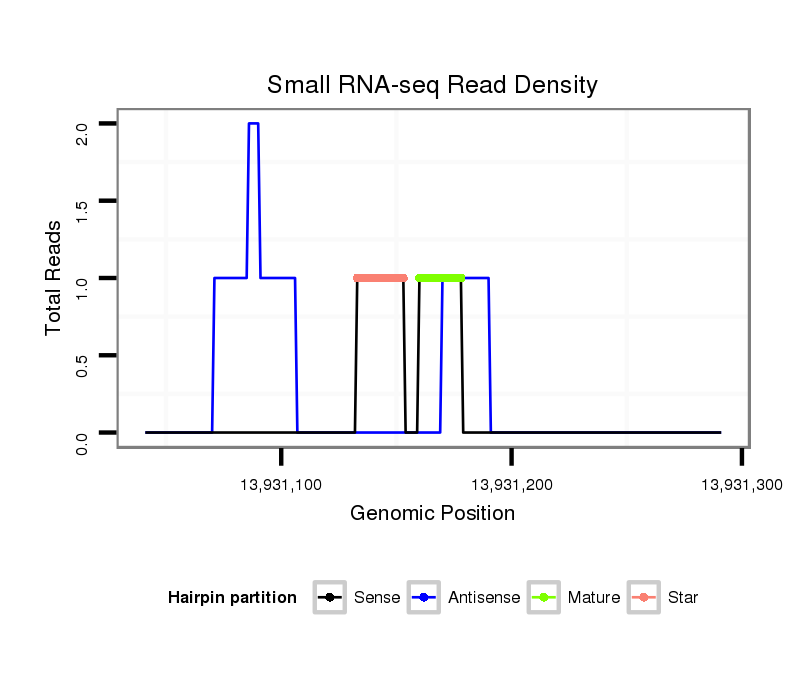
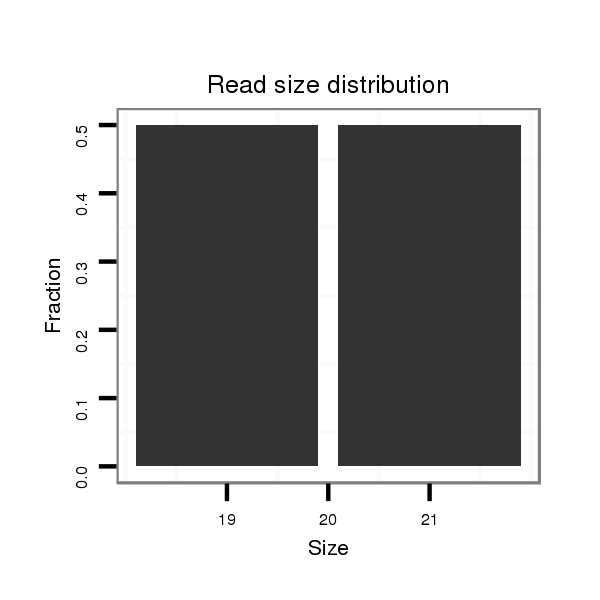
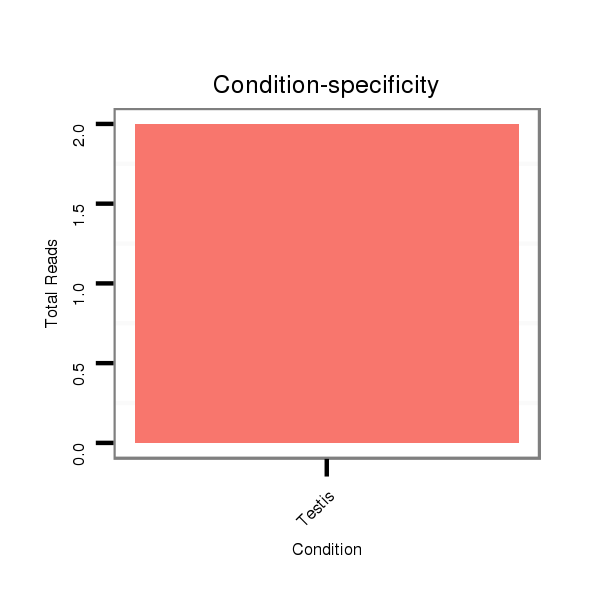
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
V053 head |
SRR060655 9x160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................GCATTCACATCACACACGC................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........AACTCCAGCCCTTGGAAATGGCGC.......................................................................................................................................................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................TTTTGGTACGCCCGTTTTGAT.......................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................CTTGGAAATGGGGGGATTAA.................................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................GTGTAATTGAGAGCAATT........................................................................................................................................................................................................ | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
|
CAATCGTCGCTGAAGTCGGGAACCTTTACCGCGCAAATTAACTCTCGTTACATTCAATGCTTTGTTTGACACGTCGAGTCGAAAACGTATTAAAAACCATGCGGGCAAAACTATATGGGCGTAAGTGTAGTGTGTGCGTATAGTGTAAACCGATCCGATAAAGTATTAAGAATTTTTATATGTAAACATACATGCGTATACACACCGACGGTAAAACCGTAGAGGGTGTTTTATTACGTTACTTTATACGC
*****************************************************************************************************************....((((..............))))....................******************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
SRR060656 9x160_ovaries_total |
SRR060671 9x160_males_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
V116 male body |
V053 head |
SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060666 160_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................GAGTAGTGTGTGCGTATA............................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ACGTCGAGTCGGAAACGTATT................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................CCATGCGGGCAAAACTAGCTAG..................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GTGTGTGCGTATAGTGTAAAC..................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GGTCGGGAACCTTTACCGCGC......................................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CGTTACATTCAATGCTTTGTT......................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GCGCAAATTAACTCTCGTTA......................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TAGGGGCAGAAGTGTAGTGT...................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................AATCCGATAAAGTAATAAG................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............AGTCCGGAACCTTTGCCTCG.......................................................................................................................................................................................................................... | 20 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ................................................................................................................ATATGGGGGTACGTGTAG......................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................ATACATGCGTGTACACTCCT............................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................CTGCGTATACAAACCTACGG........................................ | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:13931041-13931291 + | dvi_19888 | GTT--AGCAGCGACTTC-AGCCC-TTGGAAATGGCGCGTTTAATTGAGAGCAATGTAAGTTACGA--------AACA----AAC-TGTGCAGCTCAGC-----TTTTGCATAATTTTTGGTACGCCC--G-----------TTTTGATATACCCGCA----TTCA---CATCACACACGCATATCACA--TTTGGCTAGGCTATTTCATAATTCTTAAAAATATACATTTGT-------------ATGTACGCATATGTGTGGCTGCCATTTTGGCATCTCCCACA-AAATAATGCAATGAAATATGCG |
| droMoj3 | scaffold_6680:13392034-13392302 - | GTTCCAGCAGTGTCTCC-AGTCTTTTGGATATGGCGCGTCTAATTGAGAGCAATGTAAGTTACGA--------AACA----TGT---TGCAGCATAGC-----TTTTGCATAATTCTCGGTACCCCTCGG-----------GCTCGGTGTCCTCTCACA-CTTCT---CATCACGTACGCACCTCGCA--TTCAGCTAGCCTATGTCATAATTCTTAAAAATATACATTAATATGTATGAAATGTATGTATCTAAATGTGATTCCGCCA-TTTGGCATCTCCCACCCGGGGAATGCGAAGA-ATATGCG | |
| droGri2 | scaffold_15110:15309276-15309522 - | GAA--------ATCA----GGCT-TTGAACATGGCACGCCTCATCGAGAGCAATGTAAGTTACGA--------AACA----AGT---TTGAATGCAGC-----CTTTGCATAATTCCTGATAACCACCTGCCTGCCGCCCC-CTCCCTCACCCATCCCATCTTCT-CATCTATCGCACGCACATCACAATTTTGGGTAG-CTATTTCATAATTCTTAAAAATATCCATTTGT----TTTGTATGTATGT-------------------A-TTTGGCATCTCCCACA-AAAGATTGCAAAGA-ATATGCG | |
| droWil2 | scf2_1100000004762:5965755-5965862 - | GAAT---CTCCAACTTCTAGTCCCTGGAAGATGGCCCGTCTCGTTGAGAGCAATGTAAGTTACGAAATTTCA----A----AAG---TGCATATTCTCATAATTTTTATGTAATTCCCTTTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | XR_group8:4289282-4289371 - | GACC-----GTGTCT----GACTTCTGGACATGGCGCGTCTCATCGAGAGCAACGTAAGTTACGA--------CACA----ACT---TGCAGCCCAGCATCG-CTTTACATAACT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_29:523065-523151 - | C--------GTGTCT----GACTTCTGGACATGGCGCGTCTCATCGAGAGCAACGTAAGTTACGA--------CACA----AGT---TGCAGCCCAGCATCG-CTTTACATAACT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454113:2875021-2875189 - | ------------------------------ATGGCGCGTGTCGTCGAGACCAATGTAAGTTAACT----GCAAAACA----AATCCATTCACCGCTGA-----TTTTACATAACTCCCAATTCGTATCC----GCTGCCCCGCCCCCTTTTCTTTCTCA-ACTTTCT-TCTTGTTC-------TCGCA--TTCTG-----CCATTTCATAATTGTCAGCGATGATCTTTTGT----------------------------------------------------------------------------- | |
| droEle1 | scf7180000491193:3683505-3683592 + | TTA-----------------------ACAAATGGCGCGTCTCGTCGAGAGCAATGTAAGTTACTT----GC-----AATCTATTCGCTTCACTGCTGT-----TTTTACATAACTCCTGTTTCGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302334:265176-265227 + | GCA------TCGATTT----TCCGCTGGCCATGGCCCGAGTCGTCGAGAGCAATGTAAGTTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/17/2015 at 04:25 AM