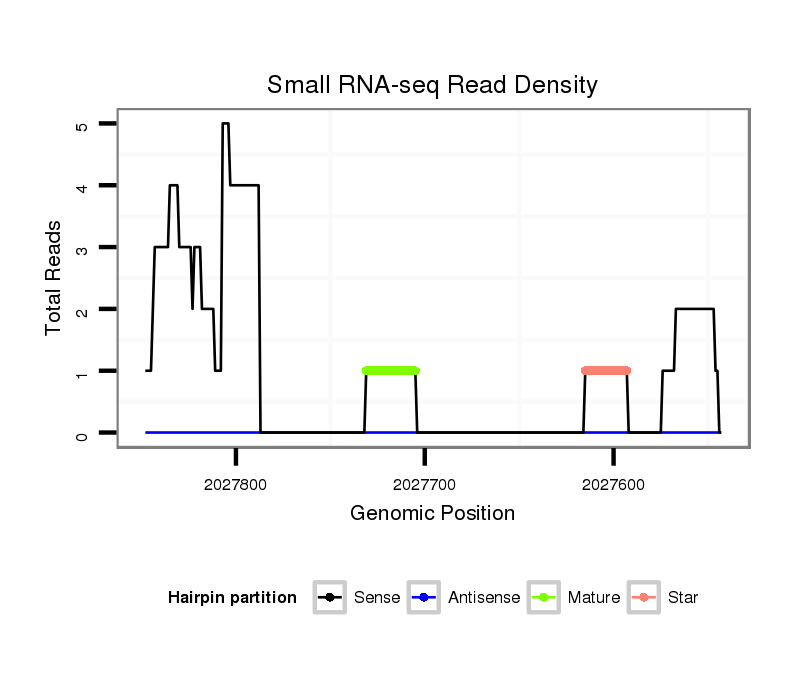
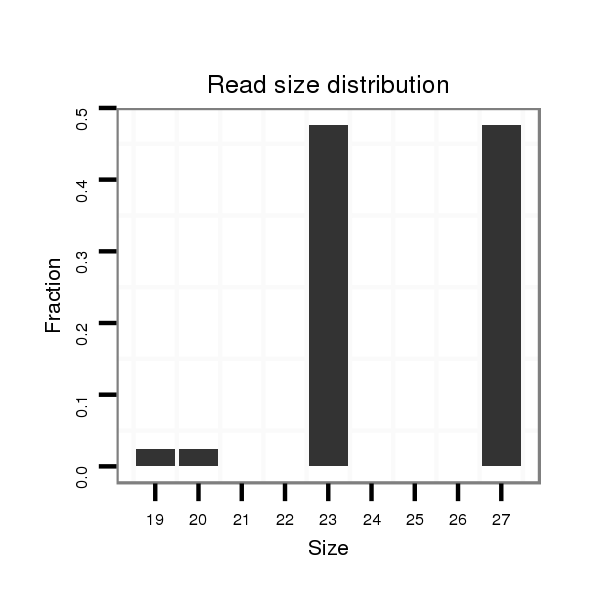
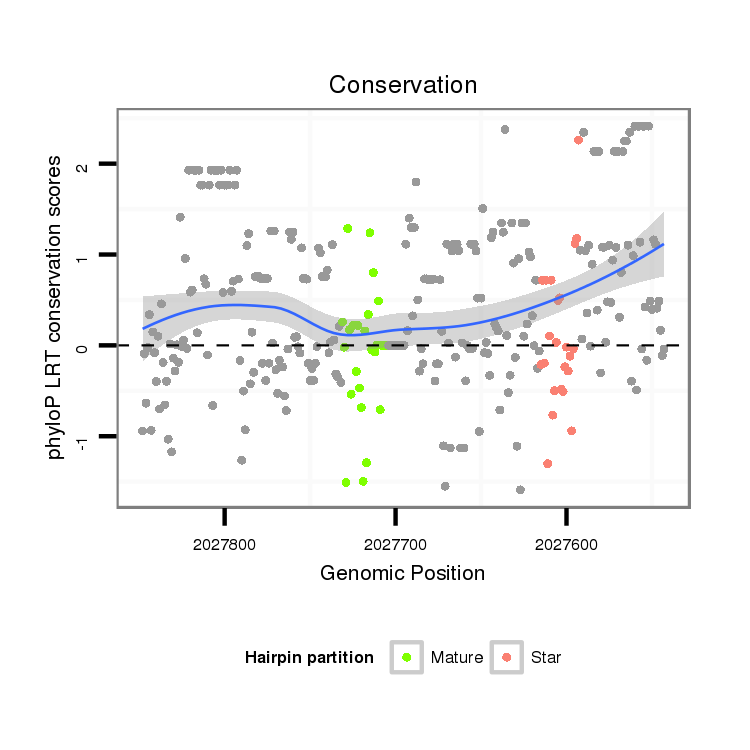
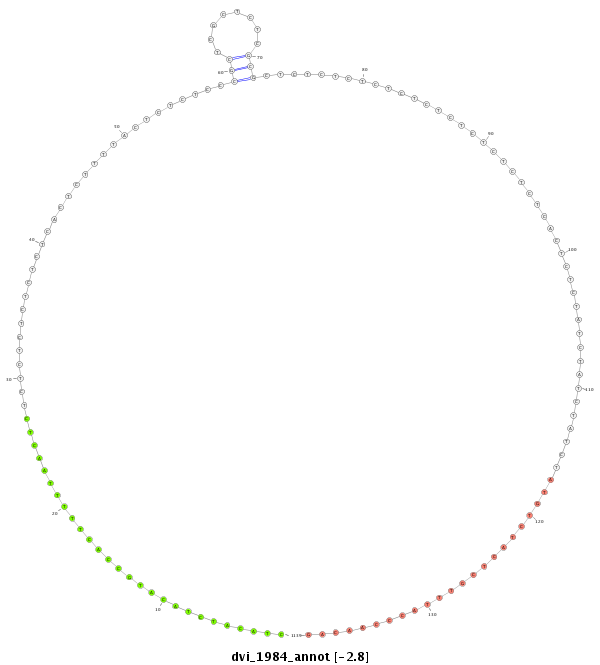
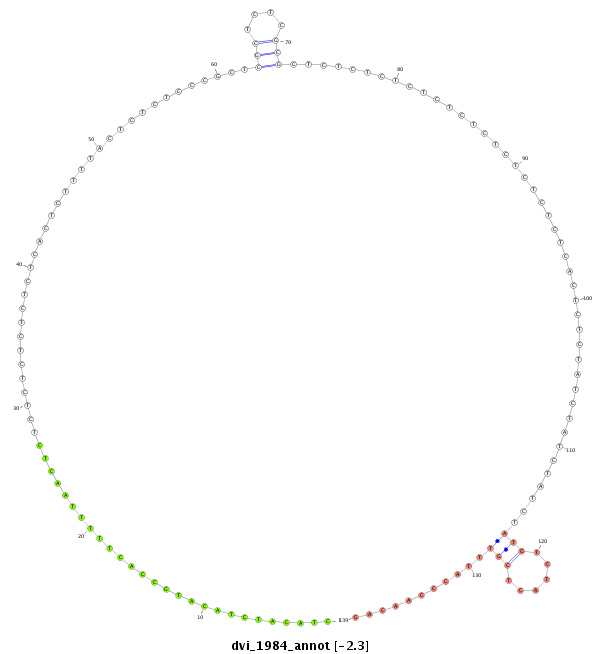
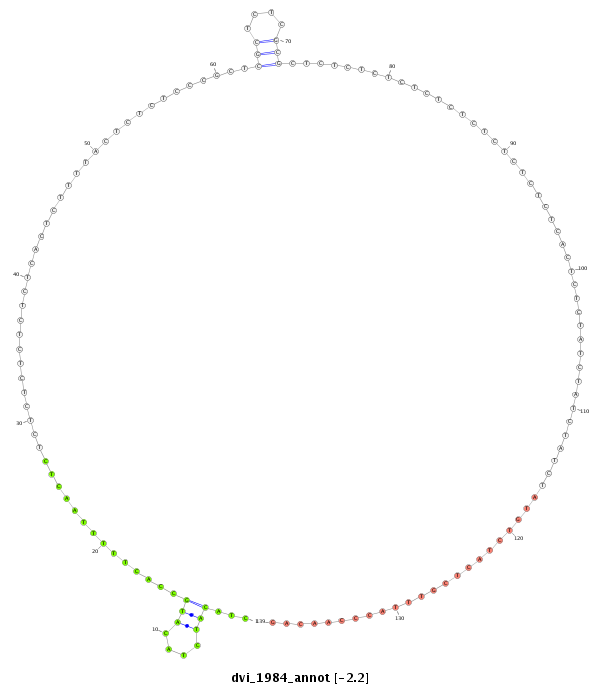
ID:dvi_1984 |
Coordinate:scaffold_12726:2027593-2027798 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -2.8 | -2.3 | -2.2 |
 |
 |
 |
exon [dvir_GLEANR_15526:5]; exon [dvir_GLEANR_15526:6]; CDS [Dvir\GJ15201-cds]; CDS [Dvir\GJ15201-cds]; intron [Dvir\GJ15201-in]
| Name | Class | Family | Strand |
| GA-rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTGCTGGCCTGTCTGACCGATTTCTTTACCAATAGCAAAGATTTTGTTCGGTGAGTTCCACGCAAAGAGTCTGAGACAAGCTCACGTCTACATTTTGTAAGACTCGAGTATCCCCGTCTACATCTACATGCCACTTTTTAACTCTCTCTCTCTCTCTCACTCTTTTACTCTCTCCCGCTCGCTCTCGCGCTCTCTCTCTCTCTCTCTCTCTCTCACTCTCTATCTATCTATCTATGTCTACTCGTTTACCCAACAGCGTTGAACGCGGCATCAAAGCTGGTGATTTTCTATTTACGTATAAGTCGA *********************************************************************************************************************..............................................................(((....)))...................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
GSM1528803 follicle cells |
M028 head |
SRR060679 140x9_testes_total |
M061 embryo |
V053 head |
SRR060672 9x160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
M027 male body |
SRR060668 160x9_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................TTTTGTTCGGTGAGTTCCAC..................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 4.00 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTACTCTCTCCC.................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............TGACCGATTTCTTTACCAATAGCA............................................................................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TTGCTGGCCTGTCTGACC................................................................................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....GGCCTGTCTGACCGATTTCT......................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................GATTTTCTATTTACGTATAAGTC.. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CTACATCTACATGCCACTTTTTAACTC.................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TGGCCTGTCTGACCGATTTCTTTACC.................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CTCAGTCTTTTACTCTCTCCA.................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TACCAATAGCAAAGATTTT..................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................AGCTGGTGATTTTCTATTTACGTATAAG.... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCATTCTTTTACTCTCTCC................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................ATGTCTACTCGTTTACCCAACAG.................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTACTCTCTCCTG................................................................................................................................. | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTACTCTCTCC................................................................................................................................... | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTCCTCTCTCCC.................................................................................................................................. | 20 | 2 | 5 | 0.40 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................ACTGCGATGAACGCGACATC.................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CTGACCGAATTCTTTAGC.................................................................................................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTTCTCTCTCCCG................................................................................................................................. | 21 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................TCACACTTTTGCTCTCTCCC.................................................................................................................................. | 20 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCACGCTTTTGCTCTCTCCC.................................................................................................................................. | 20 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCACACTTTTGCTCTCTCCT.................................................................................................................................. | 20 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTTCTCTCTCCTGCT............................................................................................................................... | 23 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTACTCTCTGCTG................................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................TTTTCAACTCTCTCTCTCTC........................................................................................................................................................ | 20 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................TCTCACATTCTCTATCTATCT............................................................................. | 21 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCACTCTTTTGCTCTCTCCT.................................................................................................................................. | 20 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAGTCTTTTGCTCTATCCC.................................................................................................................................. | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................TCTCTCTCTCTCTCTCTCTC.............................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................AACTCTCTCTCTCTCTCTC.................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TGGAACTCGGCATCAAACC............................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AACGACCGGACAGACTGGCTAAAGAAATGGTTATCGTTTCTAAAACAAGCCACTCAAGGTGCGTTTCTCAGACTCTGTTCGAGTGCAGATGTAAAACATTCTGAGCTCATAGGGGCAGATGTAGATGTACGGTGAAAAATTGAGAGAGAGAGAGAGAGTGAGAAAATGAGAGAGGGCGAGCGAGAGCGCGAGAGAGAGAGAGAGAGAGAGAGAGTGAGAGATAGATAGATAGATACAGATGAGCAAATGGGTTGTCGCAACTTGCGCCGTAGTTTCGACCACTAAAAGATAAATGCATATTCAGCT
**************************************************..............................................................(((....)))...................................................................********************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060668 160x9_males_carcasses_total |
GSM1528803 follicle cells |
M027 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
V047 embryo |
SRR060658 140_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................GGGTTGTCGCCACTTCCACC...................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................TAGCGCCCTAGTTTCGAACA......................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TGGCTAAAGAAGTGCTTAGC............................................................................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CCGGCCACTCAAGGTGCC................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................GTAAAGAAATGGTCATCG.............................................................................................................................................................................................................................................................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CTGTGCGAGTGAAGATGT...................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AGACTCGGTTCGAGTGCATG......................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................ATGGGGCCACATGTAGAT.................................................................................................................................................................................... | 18 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AGAGAGAGAGAGAGTGAGAAA............................................................................................................................................. | 21 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................GCGAGAGAGAGAGAGAGAGAGAG................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................AATGGGTTGTCGCACGTTT.......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................AGAGCGCGAGAGAGGGAG.......................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGCGAGAGAGAGAGAGAGAG.................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................GCGCGAGAGAGAGAGAGAGAGA................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................TGACAAATTGAGAGAAAG............................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:2027543-2027848 - | dvi_1984 | TTGCTGGCCTGTCTGACCGATTTCTTTACCAATAGCAAAGATTTTGTTCGGTGAGTTCCAC--------------------GCAAA--------------------------------------GAG--------TCTGAGACAAGCT---------------------CACG-------------TCTACATTTTGTAAGACTCGAGTATCCCCGTC--------------------------------------------------TA-CATCTACATGCCAC-----------------------------------------------------------------------------TTT------TT-------------AACTCTCTCTCTCTCTCTCACTCTTTTACTCTCTCCC--------------------------------------GCTCGCTCTCGCGCTCTCTCTCTC--------------------------------------------------TCTCT-----------------CTCTCTCTCACTCTCTATCTATCTA--------------------TCTATGTCTACTCGTTTACCC-------AACAGCGTTGAACGC----GGC-A-TCAAAGCTG-------------GTGATTTTCTATTTACGTAT------------AAGTCGA |
| droMoj3 | scaffold_6473:6445011-6445299 + | TTGCTGGCCTGTCTGGTCGATTTCTTTACCAACAGCAAAGATTTTGTTCGGTGAGTTTTGGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAATGTTGTGTTCGACGCACGTTTCCCAACTGCTGCGATCTCC-------CTGGATAC-GC-------------------------------------CTTTATCTCTCCCCC----CCACACACA-----CACTGT----C--------------------TCGT------------TCCCTCTCTTCCTCTCTC----------------------------------------TTTCTCTCTTTGTCTGTCACTGTCTTAC----------------TC----------G------------------C-------T----CA----------CTAT--CT----------CTCTCTATGTCTATTCGTATACCC-------AACAGCGTTGAACGC----GGA-A-TCAAAGCTG-------------GTGATTTTCTATTTACGTAT------------AAGTCGA | |
| droGri2 | scaffold_15203:994365-994725 - | TTGCTGGCCTGTCTAACCGATTTCTTTACAAACAGCGATGAATTTGTTCGGTGAGTTCGGAATTCTT-CAAAA------------------------------------------------------TATGTTGA------ACAAATGTGGGCATAGCCAAAGTTTCATATAGCCAAAACTATAATTCTGT-------------------TCTTTGCCAATGACT---------------------------------------CTATTTCCATCTACTTTGTACTAT-------TTATATTT-GT-------------------------------------ATTTTTATCTCCCTC----GCGCTCTCT-----CTCTCT----C--------------------TCTC------------TCTTT----------------------CTC--TCACTCTGTCTCTCTCTCTCTCTCTCTCTC--------------------------------------------------TCTCT-----------------CTCGCTCTCGTTCTTTATGTA---C--T--CT----------CTGTCTGTATATAATTGTAAACTC-------AACAGCGTTGAACGC----GGC-A-TCAAAGCTG-------------GTGATTTTTTATTTACCTAT------------AAGTCGA | |
| droWil2 | scf2_1100000004909:5358889-5359105 - | TTGCTGGCCTGTCTGGTTGATTTCTTTACCAATAGCAATGAATTTGTGCGGTAAGTATGAATATTTG-AAAGA--------------------------------------------------------------------------T---------------------ATAA-------------TCTATACT------------------------------------------------------------------------------------------------------------------------------ATGCTATGCTATCGCTAGTCTATCTTAA----------TCTATCT--------------------------------------------------------------------------------------------------------------------------------------ATAA----------------TCTATCACACTCACTATACCTTAC-----------TTT------------------------------CGTCTCTC-------------------------TTC-------TTCAGCGTTGAACGC----GGA-A-TTAAAGCTG-------------GTGATTTTCTATTTACCTAT------------AAGTCGA | |
| dp5 | XL_group3a:252482-252695 - | CTGCTCGCCTGTCTCACGGACTTTTTTAACAACAGCAACGAATTTGTGCGGTAAGTTTCAT--------------------GCCCT--CCATCCTT----------------------------------------------------------------------------------------------------------------------------------------A--TCCTCCGC--A------------------------------------------TTCTATAT----TCCCT-----TCTATCTTCTATGCTCTGCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCTCTATTCTCTCTTT------------------------------CGTCTCCGTT------TC----TC-CGTTCTCTTTT-------CTCAGCGTGGAACGC----GGC-A-TCAAAGCTG-------------GCGATTTCCTTTTCACGTAC------------AAGTCCA | |
| droPer2 | scaffold_39:481070-481283 + | CTGCTCGCCTGTCTCACTGACTTTTTTAACAACAGCAACGAATTTGTGCGGTAAGTTTCAT--------------------GCCCT--CCATCCTT----------------------------------------------------------------------------------------------------------------------------------------A--TCCTCCTC--A------------------------------------------GTCCAGAT----TCCCT-----TCTCTCTTCTATGCTCTGCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCTCTATTCTCTCTTT------------------------------CGTCTCCGTT------TC----TC-CGTTCTCTTTT-------CTCAGCGTGGAACGC----GGC-A-TCAAAGCTG-------------GCGATTTCCTTTTCACGTAC------------AAGTCCA | |
| droAna3 | scaffold_13258:1533001-1533205 - | TGTCTCTGTCTCTGTCTCTGTCTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTCTCTGTC------------TCTGT----------CTCTGTCTCT--GTCTCTG----------------------------TCT----------CT------------CTGTCTGTCTGTCTCTCTGTCTCTCTGTCTCTCTGT--CTCTCTG-------T----CT----------CTAC--CTCTGTCTCTGT----CT----C------TGTCTCT-------AGCAGTTTTTATAGCCAACAGAGA-CAGAGACAG-------------CCGATTTTCTATCCATTTTTATTGATATATTAAAGATGA | |
| droBip1 | scf7180000396741:81190-81370 - | AAAAAT---------------------------------------------------------------------------------------------------------------------------------TTTTAAAAATCCG---------------------CCCA-------------TCTATTTTTTGCCTTGCTC----------------------------ACTATCCTGCCAA-----------------------G-CAGCCACA-------------------------------A-----------------------------------------------CCGATCT------CT-------------A--------------------TCAC------------TGCCTCTCTATCTCTCTC----------------------------------------TCTCTCTATCTCTGTCTCGCTGTCTAACTATCTA--------------TGTC--TCTGT-----------------CTCTATCTGACT----GTCTATCTAACC--CA------------------------------------------------------------------------------------------------CTC----TCTAT------------ATATATT | |
| droKik1 | scf7180000302258:79543-79690 + | TCTCTCTCTCTCTCTACTCTCTCTCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTCTCTC------------TCTCT----------CTCTCTCTCT--CTCTCTC----------------------------TCT----------CT------------CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCT--CTCTCTC-------T----CT--C------TCTCTCTCT------CTCT----CT----C------TCTCTCGATCAACCTATCGCT-----------------------------------------------------------------------------TT | |
| droFic1 | scf7180000453829:1341146-1341294 + | ACTTAGTAGCACCCTCTATGTATC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACTT-----------------------------------------------TCGGCCCA-----CTCC-----------------------------------------------C----C------------C----------------------------------------------------------------------------------------------C--ATCTTTCCGC--ATCCCTT-------T----CT----------CTAC--TTTCTCCACT------TC----TC-TCT-TTCCCCC-------GACAGCGTTGAACGC----GGC-A-TCAAAGCTG-------------GCGATTTCCTTTTCACCTAC------------AAGTCGA | |
| droEle1 | scf7180000490214:1137165-1137346 - | TCTTTCTACCACTGTCTGCCTATC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTCTCTC------------TCTCT----------CTCTCTCTCT--CTCTCTC----------------------------TCT----------CT------------CTCTCTCTCTCTCTCTCTCTCTCTCGCTTCCTCT----CATGTTT-------T----AA--C------CCTCTCCTTCTGGCTG--------------------TCTCGTC-------TCCAGCTTTAGCCGC----AGT-CATTTTGGCTGCCAAAACTTGTGCACG-------ATTTA---------------------CGT | |
| droRho1 | scf7180000779488:477698-477823 + | CCATTATCGTGCTCTCTCTCTCTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTCTCCTCTACTCTACTCT--------------------------------------ACTCTCTGTCTCTATATATCTGTC--------------------------------------------------TATAT-----------------CTATCTCTCGTTCGTTATCTATCTA---------------------------TC-CA--------------------------------------------------------------------CTGCTCTTTATCTCC------------GAAGCGT | |
| droBia1 | scf7180000299393:9065-9207 + | TCTCTCTCTCTCTCTCTCTCTCTCT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTCTCTC------------TCTCT----------CTCTCTCTCT--CTCTCTC----------------------------TCT----------CT------------CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCT--CTCTCTC-------T----CT--C------TCTCTCTCTCTCTCTGT------CT----CT-CTCTCTTGCTT-------GAC----------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415352:1038566-1038722 + | GCTTCCGTTCCCAGTGTGTAATTTTCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCCCGTT--------------------------------------------------TC-CCTTTGTTGT--A---G-------TTGTTTATCTTGTCTC-----------------------------------------------TCGCTCTC-----TCTCTCT----CTCA------------------------------------------------------CTCTGTGTGTCTT--TGTCTCTGTTGCTCTCGTGTCCTCTCTGTC--------------------------------------------------TCACT-----------------CCCTGTCTCACTCTG-ATCTG--------------------------------------TT-----------------------------------------------------------------------------------------------TAT | |
| droEug1 | scf7180000409528:468396-468668 - | CTACTAGCCTGTCTGGTCGATTTCTTTACCAACAGCAGCGAATTTGTGCGGTAGGTTTAGTC--------------------CCC--------------------------------------------------------------------------------------------------------------------------------CTTTTAAAGCCCCATACTAA--CTCCCCACCTAGATTAGATACTAGACATACGTAG----------TAGAACCTT-------CTGTATCCCTTGTCTT-----------------------------------------------TTGGTCTA-----CTACTCT----ACCC------------------------------------------------------C----------------------------------------------------------------------------------------------CCTGTATCTCTACCTTCCCTTT-------T----CT----------CTAC--TTTCTCCTGT------AC----TT-TCTTTTCCCCC-------GACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTCCTTTTCACCTAC------------AAGTCGA | |
| dm3 | chrX:18286144-18286417 - | CTCCTTGCCTGTCTCGTCGACTTCCTAACCAACAGCAGCGACTTTGTGCGGTAAGTTTGAATCGCTTGAATGGAAAATATTGCTCCTGCTAGCTTTATAAACCTTTTGATCTTGCATTTATC--------------------------------------------------------------------------------------------------------------------------TGAAA-AGAT--------------------------------AT-------ATGTATCT-GTGCCTT-----------------------------------------------CTGGTCTA-----CTCC-----------------------------------------------C----------------------------------------------------------------------------------------------------------TGGTCTATTGCTGCTCTAT--TCCCCTT-------T----CT----------CTAC--TTTCTCCTAC------TC----AT-TCTTTTCACCC-------GACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTACTTTTTACCTAC------------AAGTCGA | |
| droSim2 | x:17383857-17384124 - | CTTCTTGCCTGTCTCGTCGACTTCCTAACCAACAGCAGCGACTTTGTGCGGTAAGTTGGAATCGTTTGAATGGAAAATCTTGCTCC--CTAGCTTTATAAACCTTTTGATCTTGCATTTGTG--------------------------------------------------------------------------------------------------------------------------TGGAA-AGAT--------------------------------AT-------CTGTATCT-GTGACTT-----------------------------------------------CTGGTCTA-----CTGC-----------------------------------------------C----C------------C----------------------------------------------------------------------------------------------GCTGCTGCTCTAT--TCCCCTT-------T----CT----------CTAC--TTTCTCCTAT------TC----AT-TCTTTTCACCC-------GACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTACTTTTTACCTAC------------AAGTCGA | |
| droSec2 | scaffold_8:595512-595779 - | CTTCTTGCCTGTCTCGTCGACTTCCTAACCAACAGCAGCGACTTTGTGCGGTAAGTTGGAATCGTTTGAATGGAAAATCTTGCTCC--CTAGCTTTATAAACCTTTTGATCTTGCATTTGTT--------------------------------------------------------------------------------------------------------------------------TGGAA-AGAG--------------------------------AT-------CTGTATCT-GTGACTT-----------------------------------------------CTGGTCTA-----CTGC-----------------------------------------------C----C------------C----------------------------------------------------------------------------------------------GCTGCTGCTCTAT--TCCCCTT-------T----CT----------CTAC--TTTCTCCTAT------TC----AT-TCTTTTCACCC-------AACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTACTTTTTACCTAC------------AAGTCGA | |
| droYak3 | X:16914229-16914533 - | CTCCTTGCCTGTCTCGTCGACTTCTTAACCAACAGCAGCGATTTTGTGCGGTAAGTTTGGATCGTTT-CATAGAACCTAATGCTCC--TAAGCTCTATAGACCTCATGATCTTGCCTTTATCGGGAGAATGTTGA------AAACC-G------------A--------C--------------------------------TTCGAGTGTCCTTGTT------T-TTGCCTTTCGATTCTGCCTAGAA-A--------------------------------------------------------ACTT-----------------------------------------------CTGATCT------CTGC-----------------------------------------------C----C------------C-----------------------------------------------------------------------------------------------CTGCTGCTCTAT--TCCCCTT-------T----CT----------CTAC--TTTCTCCTCT------TC----AT-TCTTTTCACCC-------GACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTACTTTTTACCTAC------------AAGTCGA | |
| droEre2 | scaffold_4690:8606644-8606936 - | CTTCTTGCCTGTCTCGTCGACTTTCTGACCAACAGCAGCGATTTTGTGCGGTAAGTTTGGATTGTTT-CTTAG---------------------------------------------------------------------------------------------------------------------------------------TGTCCTTGCTGAAGACT-ATGCCTA--GTTCCTACCTAGAA-AGACATTAG-CACGCGTGG----------TAGTACTGT-------CTGTATCT-GCGACTT-----------------------------------------------CTGATCTG-----CTGC-----------------------------------------------C----CCTCTGCCCCTCTG----------------------------------------CCCCTCT-----------------------------------------------GCTGCTCCTCTAT--TCCCCTT-------T----CA----------CTAC--TTTCTCCTCT------TC----GT-TCTTTTCACCC-------GACAGCGTTGAGCGC----GGC-A-TCAAAGCTG-------------GCGATTTACTTTTTACCTAC------------AAGTCGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:38 AM