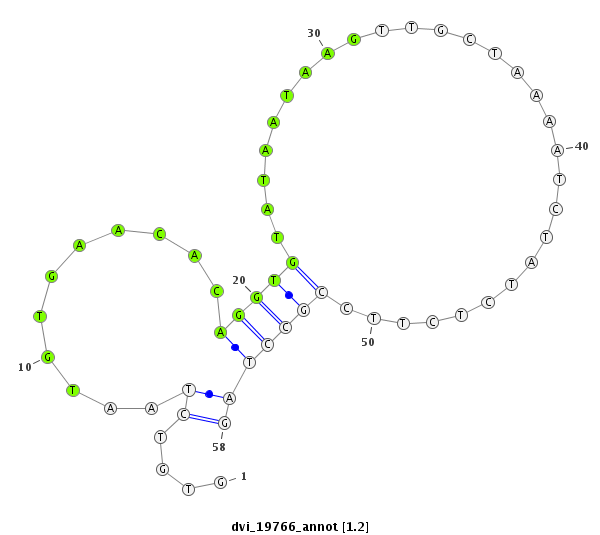
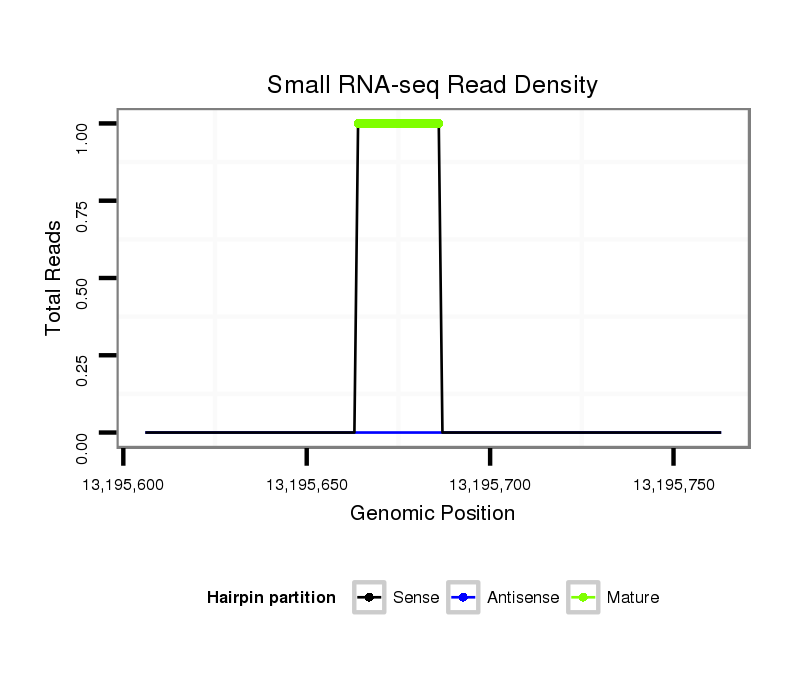
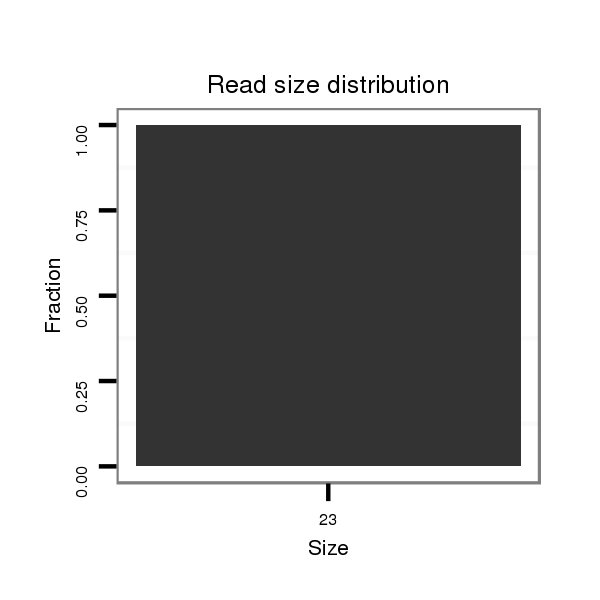
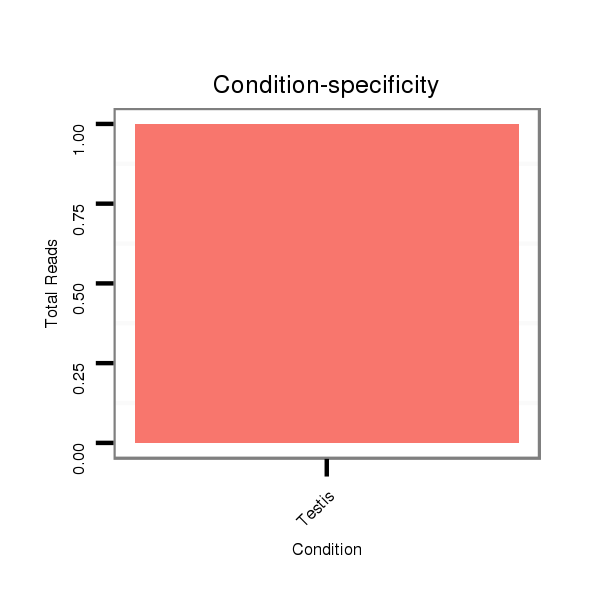
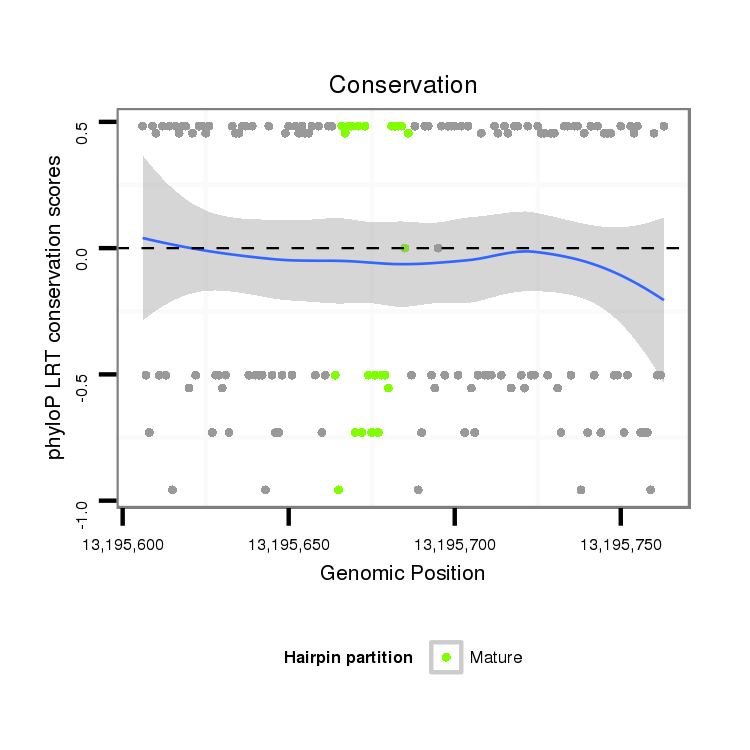
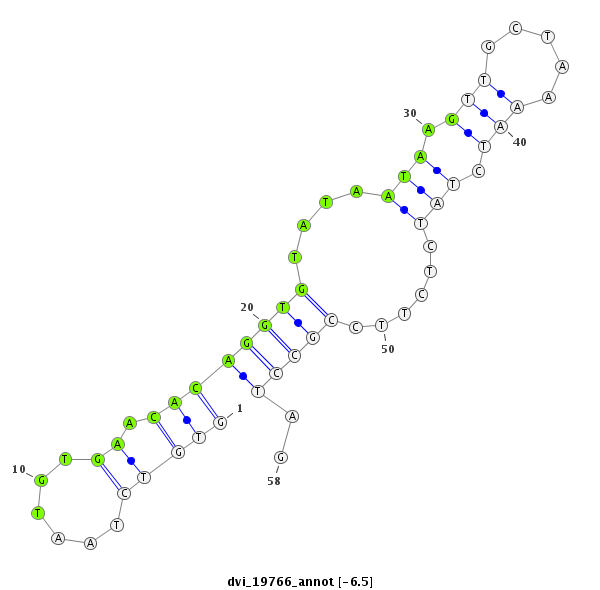
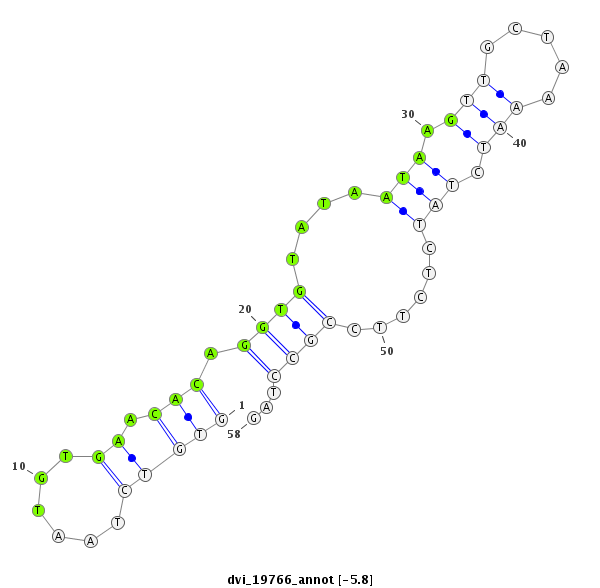
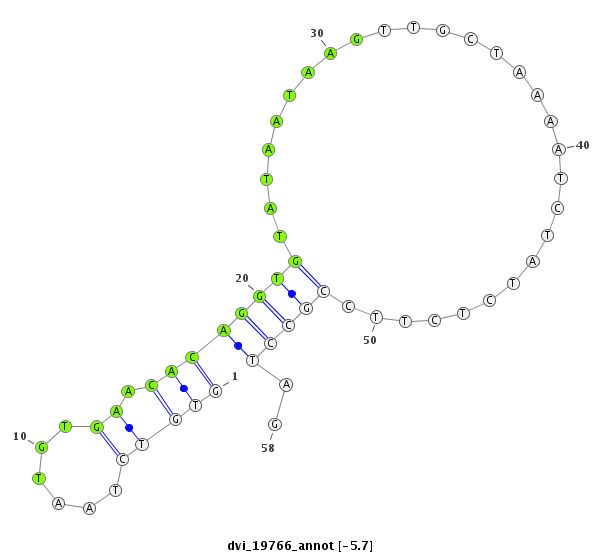
ID:dvi_19766 |
Coordinate:scaffold_13049:13195656-13195713 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -6.5 | -5.8 | -5.7 |
 |
 |
 |
exon [dvir_GLEANR_13394:2]; CDS [Dvir\GJ13507-cds]; exon [dvir_GLEANR_13394:1]; CDS [Dvir\GJ13507-cds]; intron [Dvir\GJ13507-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------################################################## AACTGCAGTCAGAATCCTTCAATATGCAGCAAGAACGCAGAGACTCTCAGGTGTCTAATGTGAACACAGGTGTATAATAAGTTGCTAAAATCTATCTCTTCCGCCTAGTTGTAAGTACCTGGGCCACAACTTGGATGACCCCAATCATCACACGGTCT **************************************************....((...........(((((.............................)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060666 160_males_carcasses_total |
M047 female body |
SRR060655 9x160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................ATGCAGCAAAAGCGCAGAGA................................................................................................................... | 20 | 2 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TGTGAACACAGGTGTATAATAAG............................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TCAGGTCTCTAAGGTGAA.............................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................AAAATGCAGCAAAAGCGCAGAG.................................................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................CTAATGTGAAAACAGGTATC.................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................ATGCAGCAAAAGCGCAGA..................................................................................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................GATGAACGGAGAGACTCTC.............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTGACGTCAGTCTTAGGAAGTTATACGTCGTTCTTGCGTCTCTGAGAGTCCACAGATTACACTTGTGTCCACATATTATTCAACGATTTTAGATAGAGAAGGCGGATCAACATTCATGGACCCGGTGTTGAACCTACTGGGGTTAGTAGTGTGCCAGA
**************************************************....((...........(((((.............................)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060678 9x140_testes_total |
SRR060663 160_0-2h_embryos_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................CGGTGTTGGACCTACAGGGT................ | 20 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................CCCGGTGTTGTAGCTACTGGGC................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................CCCGGTGTTGGACCGACTGGGC................ | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CCGGTGTTGTAGCTACTGGGC................ | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| .........................................................................................................................CCGGTGTTGCAGCTACTGGGC................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CCGGTGTTGTTCCTACTGGGA................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CCGTTGTTGATTCTACTGGGG................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CCAGTGTTGTACCTACTGGGC................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CCGGTGTAGTACCTACTGGGC................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................CGGTGTTGCAGCTACTGGGC................ | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................CGGTGTAGTACCTACTGGGT................ | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................TAGAGAGGGAGGCTCAACAT............................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................TTATTTAACGATTTTAGA................................................................. | 18 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................CGGTGTAGTAGCTACTGGGG................ | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................CGATGTTGATGCTACTGGG................. | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ....AGGTAGTCTTAGGAAGTTAT...................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................CGGTGTAGACGCTACTGGG................. | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................CTACTGGGGCTGGCAGTG....... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................GGTGTTGTAGCTACTGGGT................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:13195606-13195763 + | dvi_19766 | AACTGCAGTCAGAATCCTTCAATATGCAGCAAGAACGCAGAGACTCTCAGGTGTCTAAT----------GTGAACACAGGTGTATAATAAGTTGCTAAAATCTA-TCTCTTCCGCCTAGTTG------TAAGTACCTGGGCCACAACTTGGATGACC---CCAATCATCACACGGTCT |
| droMoj3 | scaffold_6680:14167583-14167758 - | AGATGTAATGAGAAACTTTCACCGAAAAGCAAAAGTAGAACTGCTTTCAGGTATACAACAAATTGTAGTCTGAAAAAAATCTCGAAATA-GCTCATAGT-TTTATTTTATAATGTTCAGCTGTAGTGAAAAAAATTTGGACCTAAATTTCGCTAAACCGTCCGGTAGTCAACACGCTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:31 PM