
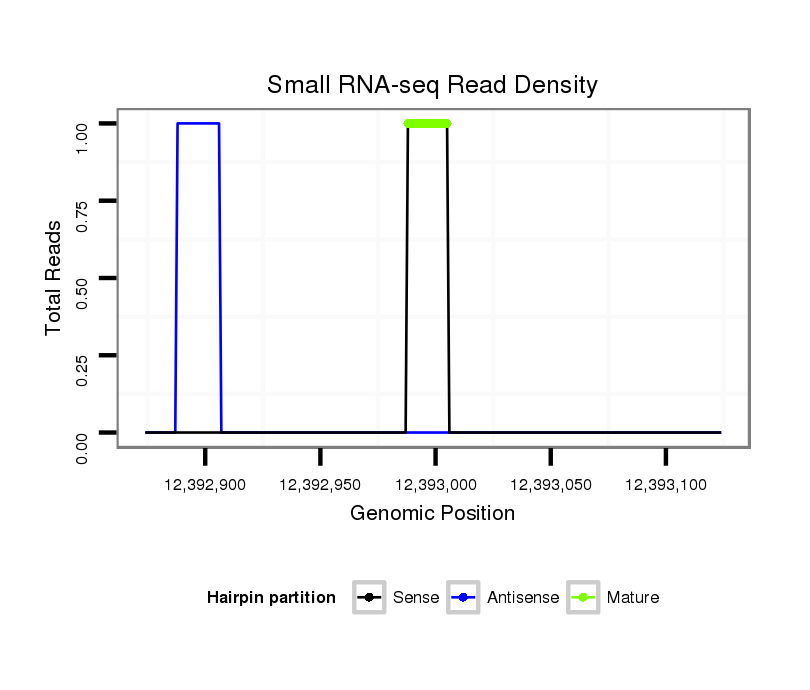
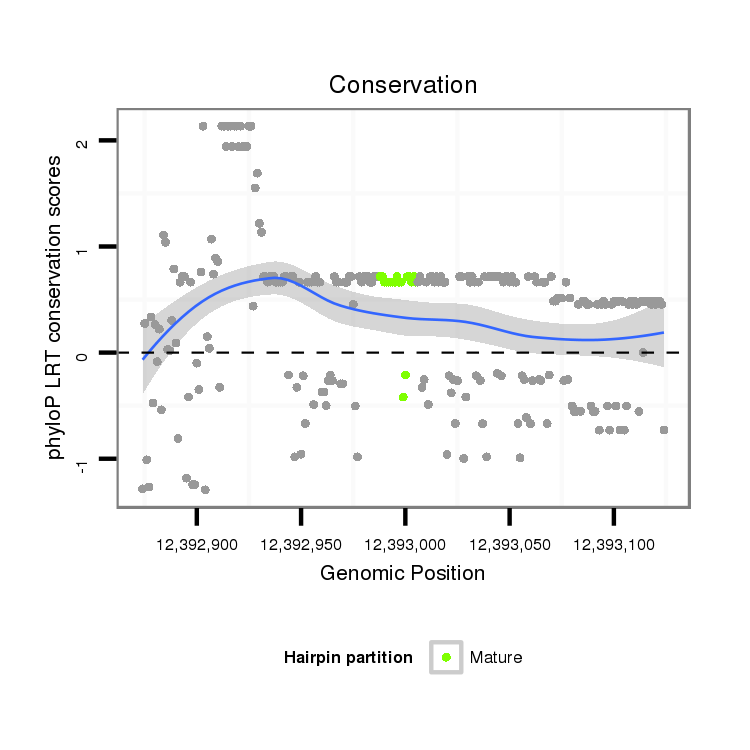
ID:dvi_19672 |
Coordinate:scaffold_13049:12392924-12393074 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
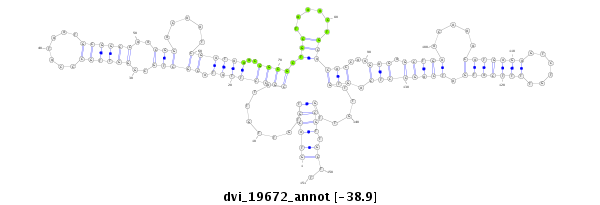
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -38.9 | -38.9 | -38.9 |
 |
 |
 |
CDS [Dvir\GJ13461-cds]; exon [dvir_GLEANR_13350:1]; intron [Dvir\GJ13461-in]
No Repeatable elements found
| --------------------------------------############--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACGTAACGCTAGCGAGTCCAAAAGCGTTCATCTTCAAAATGATGTTCACGGTACGTTGTTGTTACGCCTTATACGCTGCGGGTTGCCCATAATGCGGCCAACGGACAATCGATAAACGGCGCTGCTAAACTGCACACCAAGAGAGCTGAACAAACATGAGAGTCTGTTTTCATGATCAGCCTCAGTGTTGTTGCGTTGCTTATGTTGTTCTATGCCATCTCTCCGCTCTCTGCGTCTCATTTTACTCAAGC **************************************************(((.(((...((..((((((((.(((((..((((((.......))))))..))).....))))))..))))..))..))))))((((...(((.(((((.....((((((((....)))))))).)))))))).)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060689 160x9_testes_total |
V053 head |
M028 head |
SRR060676 9xArg_ovaries_total |
M047 female body |
SRR060659 Argentina_testes_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................AACGGCGCTGCTAAACTG....................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................CTGCGTCTTATTTTACTCAAG. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GTTCTTGCGTTGGTTATGTT............................................. | 20 | 2 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CGTCAGTGTTGTTCCGTTGTTT.................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTACGTCGATGTTACGAC....................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GCTCTCTGCGTCTCATCGT........ | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TCAGCTCCCTGCGTCTCTTT.......... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........TAGCGCGTCCGACAGCGTT............................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..GTAACGTTAGGGAGTCCG....................................................................................................................................................................................................................................... | 18 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................GTGTTCTTTGCCATGTCTCC........................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGCATTGCGATCGCTCAGGTTTTCGCAAGTAGAAGTTTTACTACAAGTGCCATGCAACAACAATGCGGAATATGCGACGCCCAACGGGTATTACGCCGGTTGCCTGTTAGCTATTTGCCGCGACGATTTGACGTGTGGTTCTCTCGACTTGTTTGTACTCTCAGACAAAAGTACTAGTCGGAGTCACAACAACGCAACGAATACAACAAGATACGGTAGAGAGGCGAGAGACGCAGAGTAAAATGAGTTCG
**************************************************(((.(((...((..((((((((.(((((..((((((.......))))))..))).....))))))..))))..))..))))))((((...(((.(((((.....((((((((....)))))))).)))))))).)))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060656 9x160_ovaries_total |
SRR060659 Argentina_testes_total |
V116 male body |
SRR060687 9_0-2h_embryos_total |
M047 female body |
SRR060682 9x140_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........ATCGCTCAGGTTTTCGCGAGT............................................................................................................................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TCAGGTTTTCGCAAGTAGA.......................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TGAAGTGTGGTTCGCTCGAGT...................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACGACTACAACAATATAGGGTA................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................AGGTTCTTTCGACTTGGTTGT............................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................AGCCCTGCAACAACAGTGCG........................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................CGGTTGAATGTTAGCTAT......................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................AGTACTAGTCAGAGGCACG............................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:12392874-12393124 + | dvi_19672 | ACGT-------AA--C-G---CTAGCG---------AGTCCAAAAGCGT-TCATCTTCAAAATGATGTTCACGGTACGTTGTTGTTACGCCTTAT------------------ACGCTGCGGGTTGCCCAT------------------AATGCG--G-CCAACGGACAATCGATAAA---CGGCGCTGCTAAACTGCACACCA----------AGAGAGCTGAACAAACATGAGAGT------C--TGTTTTCATGATCAGCCTCAGTG-TTGT--TGCGTTGCTTATGTTGTTCTATGCCATCTCTCCGCTCTCTGCGTCTCATTTTA--CTCAAGC |
| droMoj3 | scaffold_6680:15075449-15075728 - | ATTT-------AA--C-G---CTAGCGTG-------TGTGCAACTGATT-TCAGCCTCAAAATGATGTTCACGGTACGTTGTTGTTACGCCTTATGTTCGTTT----------ATTCCAAGGGGTGCTTCA------------------ATTGCGCTGCCCAACAAACAATCGATAAA---CGGCGCTGCAAAACTGCAAGCCACAAGAGAAAAAGAGAGCAAAGATAAATTGAGAGAGTACCTACATATTTTCATGATCAGCAATGCTTTATGCTGAGCTTT-----AGC-AAACAATGCTTACGCTCTTCTTTATTTGTCTCTT-TTACTCTCAAGA | |
| droGri2 | scaffold_15110:22977358-22977618 + | ACGT-------AA--C-G---CTAGCTA----GCTGAGTGCAAGTGGCTCTCAT-CTCAAAATGATGTTCACGGTACGTTGTTGTTACGCCTTGTGCTGAGTTCTGCTGAGTTAAGCAGAGGGGTGCTTATCATACATATACAGCAGCCGATGCT--T-CCAA--TACAATCGATAAAAGGCGGCGCTGCAGAACTGCACACGA--AGAAGAAGAGAGAGCCGCAAAAATTTGAGTGT------ATTTTTTTTTAAGATCAGCAGCAGTTCTTGTTATGCTCTGCTTATGTT----------------------------------------------- | |
| dp5 | XR_group6:5301302-5301351 + | CCGA-------AAA---CAGACA---------GCAAAAT---------C-ACAGTCTAACGATGATGTTCACGGTAAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_9:3611564-3611613 + | CCGA-------AAA---CAGACA---------GCAAAAT---------C-ACAGTCTAGCGATGATGTTCACGGTAAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13337:11805255-11805290 - | ACGA-------AA-----------------------------------A-GAAGC--CAAAATGATGTTCACGGTAAGTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBip1 | scf7180000396589:2175308-2175356 + | TCGG-------AGA---GTACCA---------ACG-AAA---------A-GAAGC--CAAGATGATGTTCACGGTAAGTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000302441:2269129-2269178 + | CCCT-------CC--C-G---ATAGAGACTCAGCG-AAA---------C-GCATC--CAAGATGATGTTCACGGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454048:705894-705948 - | ATAATTCCCGCAGGAGACTCACA---------GCGATAA---------G-GCATC--CAAAATGATGTTCACGGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000491255:1430878-1430919 + | GCAG-------GA------GACA---------AATAAAT---------C-TCACA--CAAAATGATGTTCACGGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000780171:15046-15090 - | CCCG------CAGG----AGACT---------AAGTAAT---------C-CCATA--CAAAATGATGTTCACGGTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 3L:13111427-13111477 + | CCGC-------AGGACTCTAATC---------ACGAAAA---------C-GCAGC--CAAGATGATGTTCACGGTAAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/16/2015 at 09:07 PM