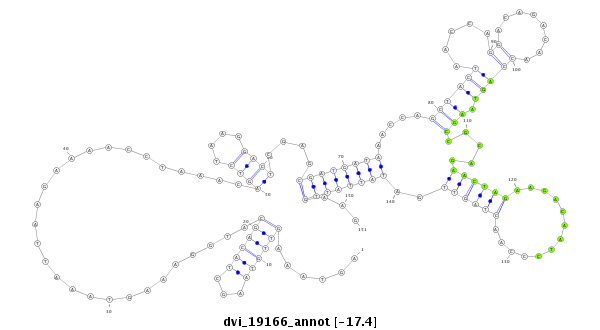
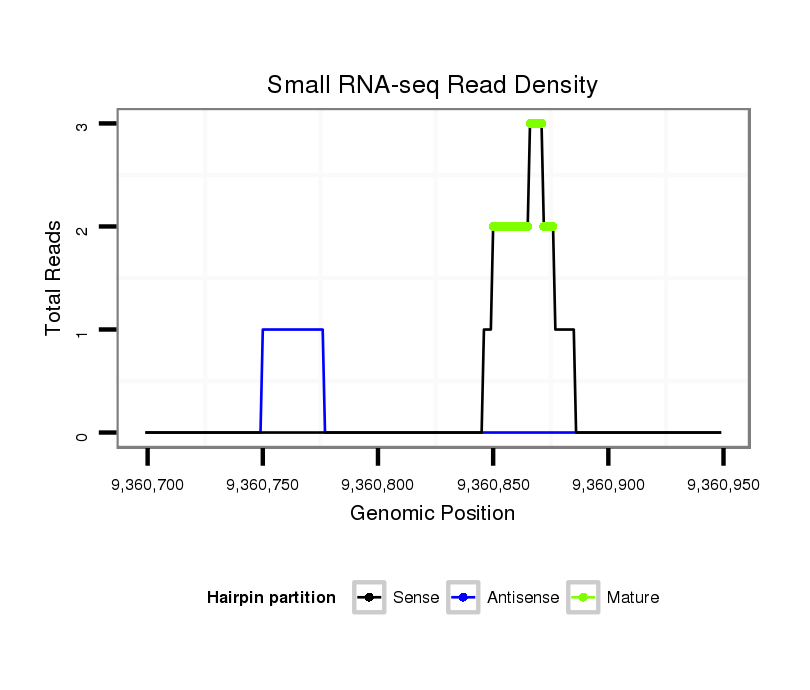
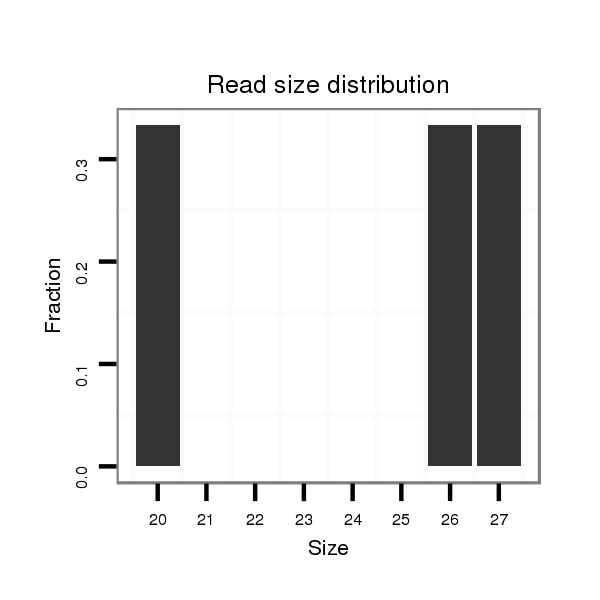
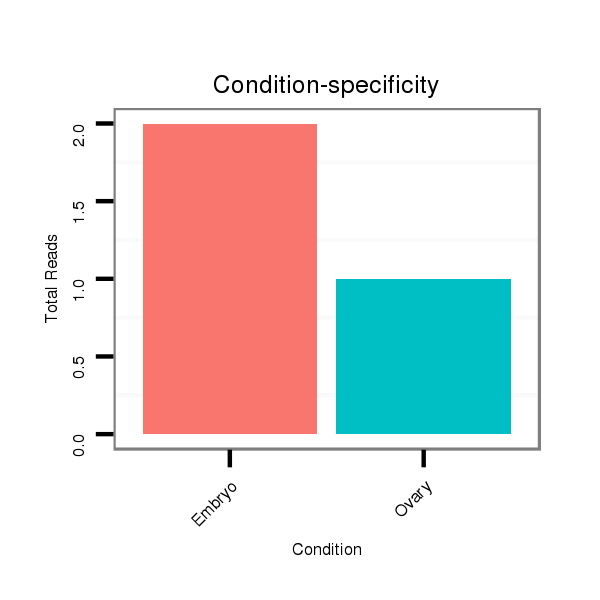
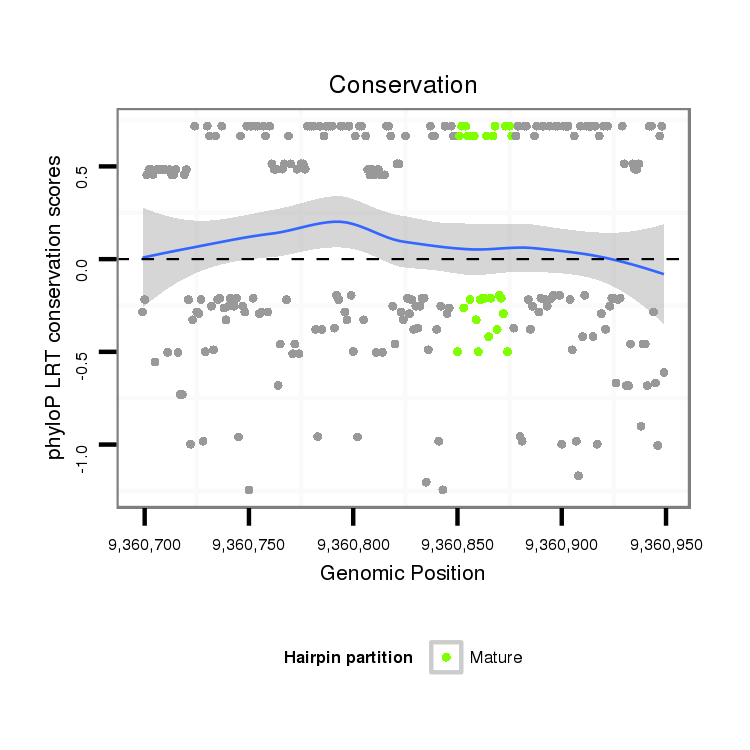
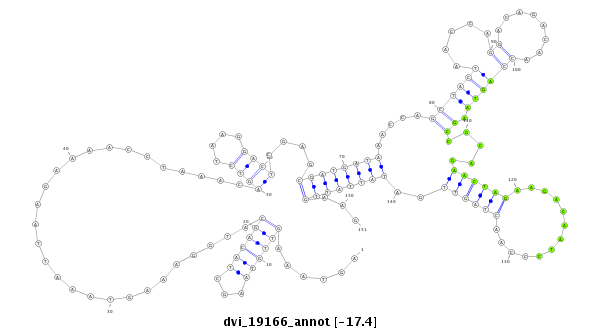
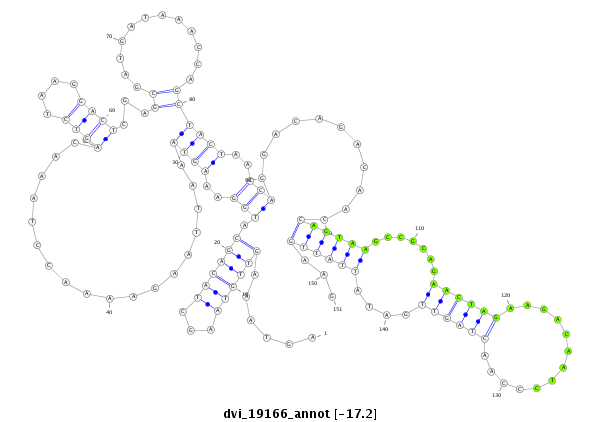
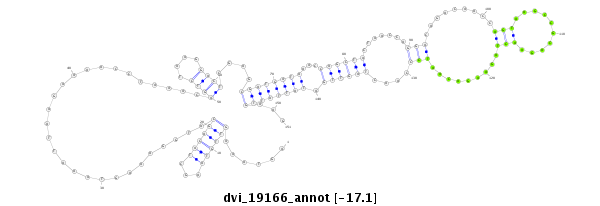
ID:dvi_19166 |
Coordinate:scaffold_13049:9360749-9360899 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -17.4 | -17.2 | -17.1 |
 |
 |
 |
exon [dvir_GLEANR_13174:6]; CDS [Dvir\GJ13272-cds]; intron [Dvir\GJ13272-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATGAAGATAAAATTGCAACCGAGCCTAGGCAAGAGCAAAAGAAGAATGATAGTAAAGTTGTAAGCTACAGCATGGAAAGTAAATTAAGAAAACCTAAACAGTCTAAGGACTCGAGCGATGATAAACCAGCTACTAACCAGGACAGACAACCAGTAAGCCGCAGAACTAGAAGACAATCCCAACTAGTTGATATTATTGAAGCAAATGGCAATGAAAATCCCAAAAATCAAAAGCATGCTGCCAAAACTGAC **************************************************......((((((...))))))............................((((....))))....((((((((.....((((((.....((........))))).))).....((((((.............))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
M027 male body |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................AGTAAGCCGCAGAACTAGAAGACAATC......................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................AGAAGACAATCCCAACTAGT................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AACCAGTAAGCCGCAGAACTAGAAGA.............................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TAGTTGACAATATTGAAGCA................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................CCGCTAGAAACCAGGACAG.......................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................CGCTAGAAACCAGGACAGA......................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................GCAGAACTAGATGACAAGG......................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| GTGAAGATAAAATCGCAGCC....................................................................................................................................................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................AACTTGAAAGCTACAGCA................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TACTTCTATTTTAACGTTGGCTCGGATCCGTTCTCGTTTTCTTCTTACTATCATTTCAACATTCGATGTCGTACCTTTCATTTAATTCTTTTGGATTTGTCAGATTCCTGAGCTCGCTACTATTTGGTCGATGATTGGTCCTGTCTGTTGGTCATTCGGCGTCTTGATCTTCTGTTAGGGTTGATCAACTATAATAACTTCGTTTACCGTTACTTTTAGGGTTTTTAGTTTTCGTACGACGGTTTTGACTG
**************************************************......((((((...))))))............................((((....))))....((((((((.....((((((.....((........))))).))).....((((((.............))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060666 160_males_carcasses_total |
M047 female body |
V053 head |
SRR060665 9_females_carcasses_total |
SRR1106721 embryo_10-12h |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CGTCTTGATGTTCTGTTA.......................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CATTTCAACATTCGATGTCGTACCTTT............................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TGGATTGGTCAGATTCCTC............................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TGATTAACTATAAGCACTTCGTT............................................... | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CTACTAGTTGGACGATGAT.................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GCTACTTTTTGGTCCACGAT.................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................CCTTTCGTTTAATCCTTTTG.............................................................................................................................................................. | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................TTTATGGTTTTTAGTTTGCGT................ | 21 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................CGTTAGTTTTAGGGTTGTTCGT...................... | 22 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:9360699-9360949 + | dvi_19166 | ATGAAGATAAAATTGCAACCGAGCCTAGGCAAGAGCAAAAGAAGAATGATAGTAAAGTTGTAAGCTACAGCATGGAAAGTAAATTAAGAAAACCTAAACAGTCTAAGGACTCGAGCGATGATAAACCAGCTACTAACCAGGACAGACAACCAGTAAGCCGCAGAACTAGAAGACAATCCCAACTAGTTGATA---TTATTGAAGCAAATGGCAATGAAAATCCCAAAAATCAAAAGCATGCTGCCAAAACTGAC |
| droMoj3 | scaffold_6680:11397004-11397227 - | GTGAAGTTAAAACTGCAGAAGAAAATGGATCAGGGCAAATTGAAGAAGGCATTAAAGCTGCA-------AC--------TAAATCAAGAAAATCTAAGAAGGCAAATGACTCAAGTGATGG---GACAGTTGTCAATCAGGATGTACTACCCGTTAGTCGACAAACAAGAAGACACTCTCAGTTAATCGACAATATTGTTGAAGAAAATGGTGAAGAAATTACCAAAGATAAAA------------AAGAGGAG | |
| droGri2 | scaffold_15110:6800426-6800650 - | GA--------------------GTCTGTGAAAGACCTGAAGAGGAGCGACACTGAATCTGCAAGCGGCAGCAAAGTAAGTAAAGAACGAAAATTAAAGCAATCCAAGG---------ATGACAAGCCGTCGATTGGGGAGGCAACAAAACCAGTAAGCCGCAGGGCAGGACAGAAATCTCATATAGGTGACGTTGTAATCAAAATAAAACGACAAAAAATTTCAACAAGCATGAATAGTGCTCTTCAAGAAGAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 09:04 PM