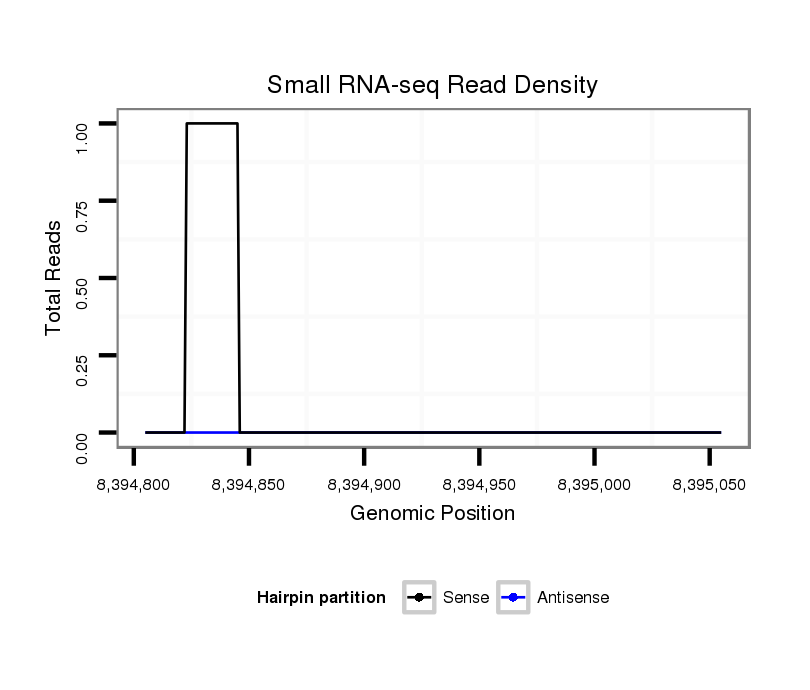
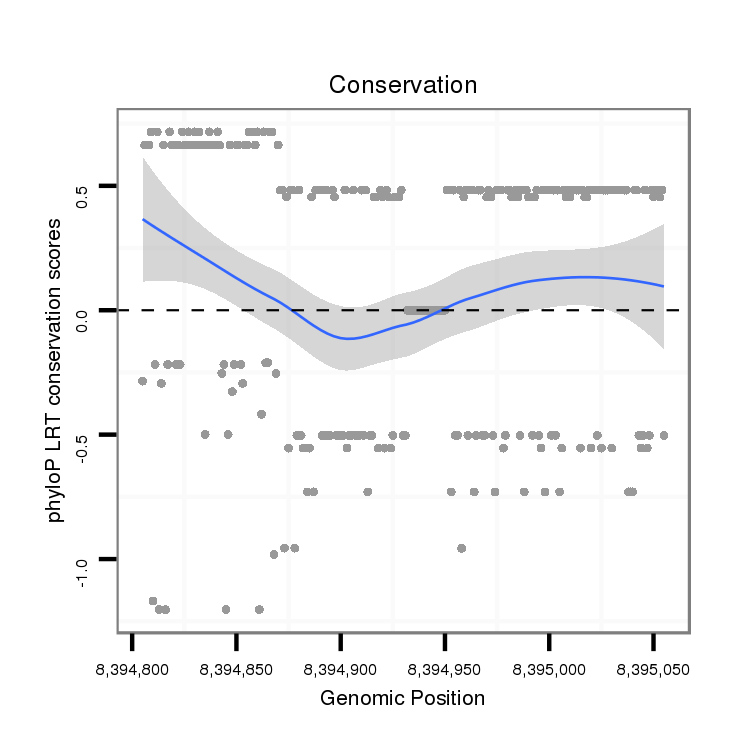
ID:dvi_18984 |
Coordinate:scaffold_13049:8394855-8395005 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ13197-cds]; exon [dvir_GLEANR_13106:1]; intron [Dvir\GJ13197-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACGCTTCTCGCCCTGGCGCTGGAGCAGAGCAGTCGCTGTGCAGCCGCCGCGTAAGTCTATTAACTGAACCATACCTGTTGTCCAAACCAGTTGGGATTATGTCCAAGTGCGCCTTGATCTTGGCTTGATTGGGGATTTGCAGTGAGATCTCAAGCTCAAGCTTACGTGCCAAAATATGCTGTTATGCACACAACATCTCTCAAGTCATAATTGCATTTATATAATATAAATTTCAAAAATTAAAACTTGTC **************************************************((((((..................((((.((((...(((((((((.......(((((..((...))..))))))))))))))))))..))))((((........)))).))))))((((...((((...))))..))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
M047 female body |
SRR060658 140_ovaries_total |
V116 male body |
M028 head |
SRR060687 9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................CTGGAGCAGAGCAGTCGCTGTGC.................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........CCTGGCGCTGGAGCAGATCC............................................................................................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TCTGAACCCTACCTGTTGTGCA....................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GATCCGGGCTTGATTGGGAAT................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TGGGGAGGTCCAGTGAGAT....................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........TCCTGGGGCTGGAGCAGGGCA............................................................................................................................................................................................................................ | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TCTGTGCATGATTGGGGATT.................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........CCTGGGGCTGGAGCAGGGC............................................................................................................................................................................................................................. | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........CCTGGGGCTGGAGCAG................................................................................................................................................................................................................................ | 16 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....TCACGTCCTGGGGCTGGA.................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................AAGTGAGCCGTACCTGTTG........................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TGCGAAGAGCGGGACCGCGACCTCGTCTCGTCAGCGACACGTCGGCGGCGCATTCAGATAATTGACTTGGTATGGACAACAGGTTTGGTCAACCCTAATACAGGTTCACGCGGAACTAGAACCGAACTAACCCCTAAACGTCACTCTAGAGTTCGAGTTCGAATGCACGGTTTTATACGACAATACGTGTGTTGTAGAGAGTTCAGTATTAACGTAAATATATTATATTTAAAGTTTTTAATTTTGAACAG
**************************************************((((((..................((((.((((...(((((((((.......(((((..((...))..))))))))))))))))))..))))((((........)))).))))))((((...((((...))))..))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060670 9_testes_total |
SRR060660 Argentina_ovaries_total |
M047 female body |
V116 male body |
SRR060676 9xArg_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060655 9x160_testes_total |
SRR060658 140_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................GATGTGGAGAGTTCAGTA........................................... | 18 | 2 | 4 | 1.75 | 7 | 1 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CCGAGCTAACCCCTAAACGTC............................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................GTTGTGGAGAGTTCAGTAA.......................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GGATGTGGAGAGTTCAGTA........................................... | 19 | 3 | 10 | 0.70 | 7 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| ..........................................................................................................CACGCGGAAATAGAAACGAA............................................................................................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATGTCTAGAGTTCGAGTTCGAA........................................................................................ | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TATGGGGAACAGGCTTGGTC................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GGAGAACAGGCATGGTCAA............................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................GGGGAACAGGCTTGGTCAA............................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................TCACGGTTTTGGACGACAAT................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................AGTAGGAATGCACTGTTTTA............................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:8394805-8395055 + | dvi_18984 | ACGCTTCTCGCCCTGGCGCTGGAGCAGAGCAGTCGCTGTGCAGCCGCCGCGTAAGTCTAT-------TAACTGAACCATACCTGTTGTCCAAACCAGTTGGGATTATGTCCAAGT---GCGCCTTGATCTTGGCTTGATTGGGGATTTGCAGTGAGATCTCAAGCTCAAGCTTACGTGCCAA---AATATGCTGTT---ATGC----ACACAACATCTCTCAAGTCATAAT-TGCATTTAT-----ATAATATAAATTTCAAAAATTAAAACTTGTC |
| droMoj3 | scaffold_6680:12049154-12049411 + | GCGCTCTTTGCTTTGGTGTTGGAGCAGAGCCGTCGCTGCATCGATGCTGCGTAAGTTAATCTGGCATTAATCGAAGCTTAGTTAAATACAAAATTAACTGAAGCTTCATTTGAATTATTTACCATGTTCACGGCTCA-------------------ATATTGACCTTAATTTTGTGTGTAAACGAATCATGCTGCTTATCTGCAGCTGCATTAAATTTTTATAGTCATAAAGTGCAATTGTTGTTATTAATTTAAATTTACCAAGACATGACTTGTT | |
| droGri2 | scaffold_15110:7423769-7423834 + | GCGCTGCTGTCGCTGGCGCTGGAGCAGAGCAGTCGCTGTGGAGCCGCCTCGTAAGTGAAC-------CAAATG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 07:46 PM