
ID:dvi_18968 |
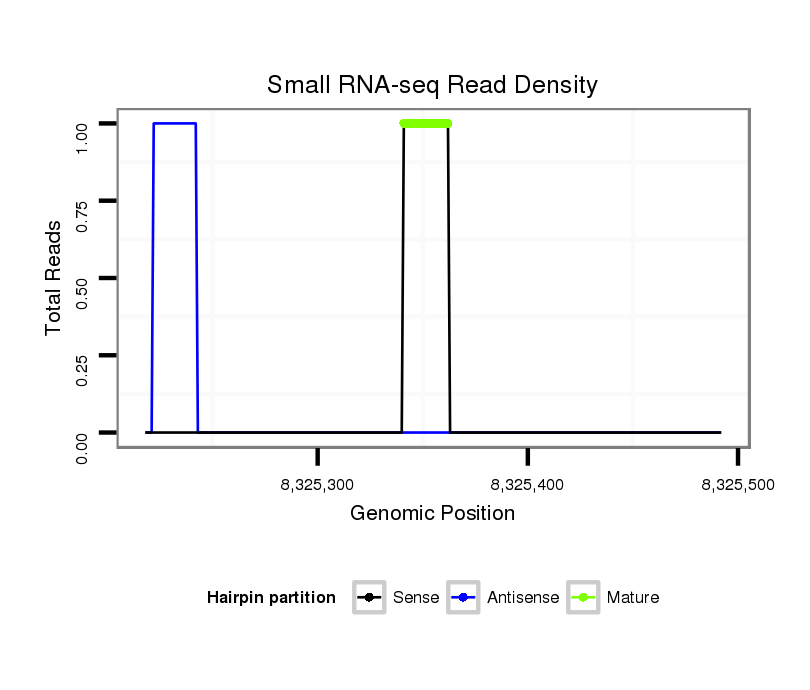
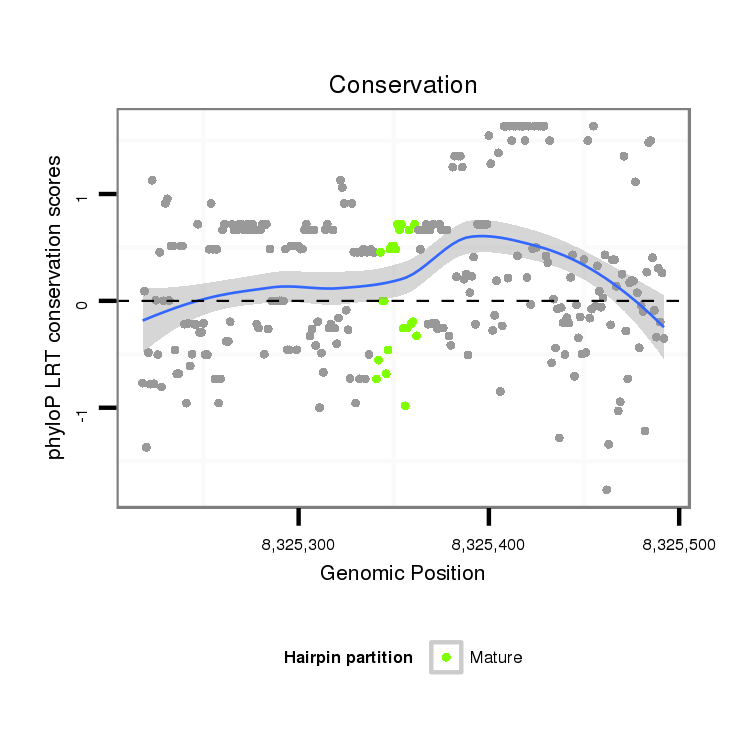
Coordinate:scaffold_13049:8325268-8325442 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -45.2 | -45.0 | -44.9 |
 |
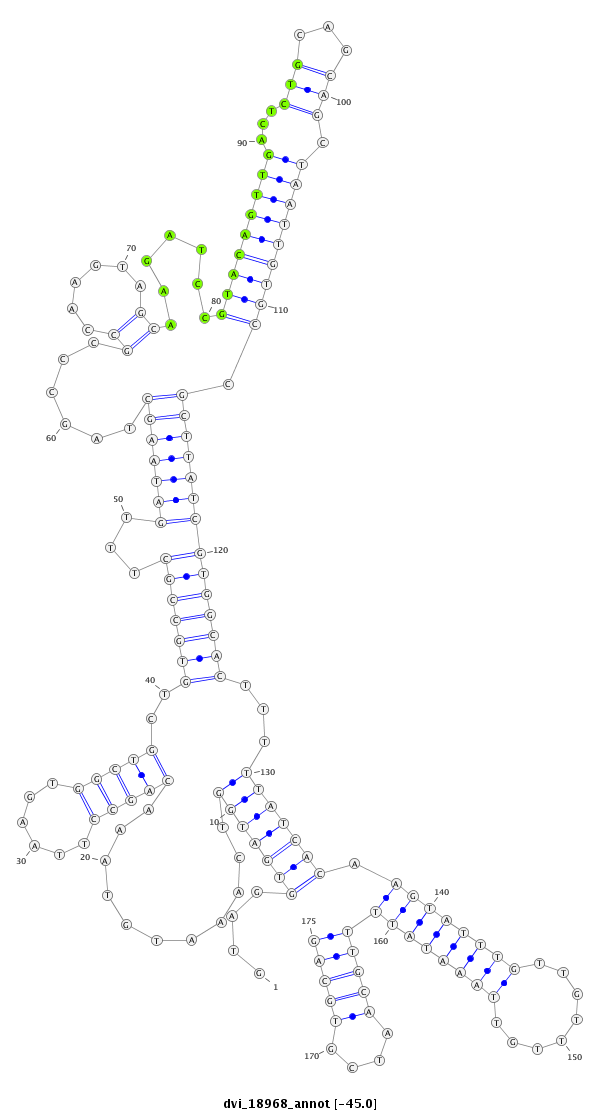 |
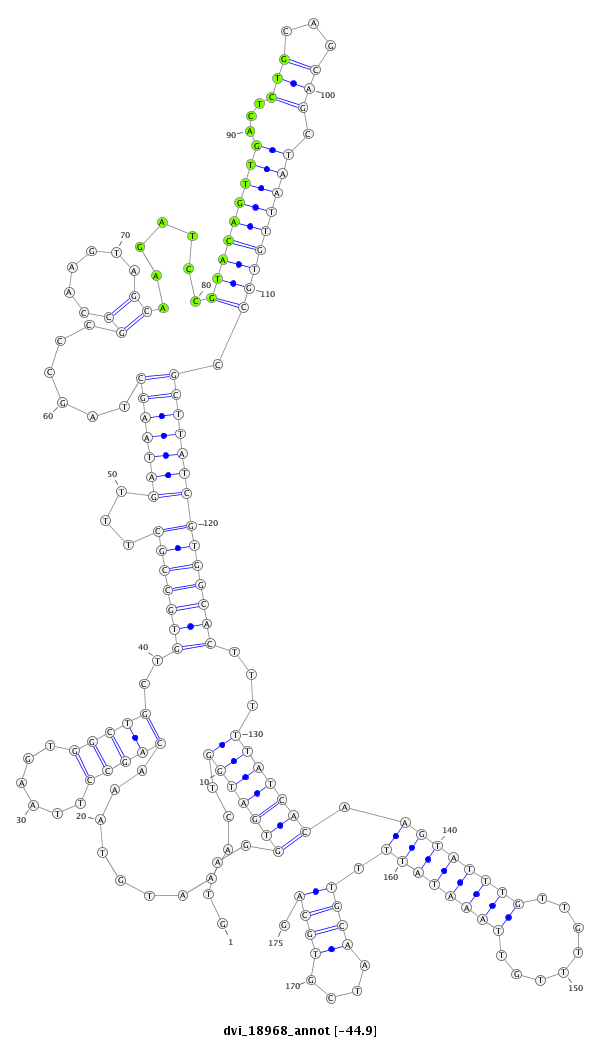 |
exon [dvir_GLEANR_13103:1]; CDS [Dvir\GJ13194-cds]; exon [dvir_GLEANR_13103:2]; CDS [Dvir\GJ13194-cds]; intron [Dvir\GJ13194-in]
No Repeatable elements found
| ----------------------------######################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGTGGAGTGCGGGATTACGCTTGATGAGATGGAGATCACTGCGAATGTGGGTAGGTGATGGTCAAATGTAAACAGCCTTAAGTGGCTGCTGTGCCGCTTTGATAAGCTAGCCCGCCAAGTAGCAAGATCCGTACAGTTGACTCTGCAGCAGCTAATTGTGCCGCTTATCGTGGCACTTTTTATCACAAGTATTTGTTGTTTGTTAAATATTTTGCAATCGTGCAGTGCATGTGCCTGTGGGCGGCAACCTTAATACCAGCTGCATATCCACATCG **************************************************....(((((((...........(((((......)))))..(((((((...(((((((......((......)).......(((((((((...(((...))).))))))))).))))))))))))))...)))))))(((((((((........)))))))))((((....)))).************************************************** |
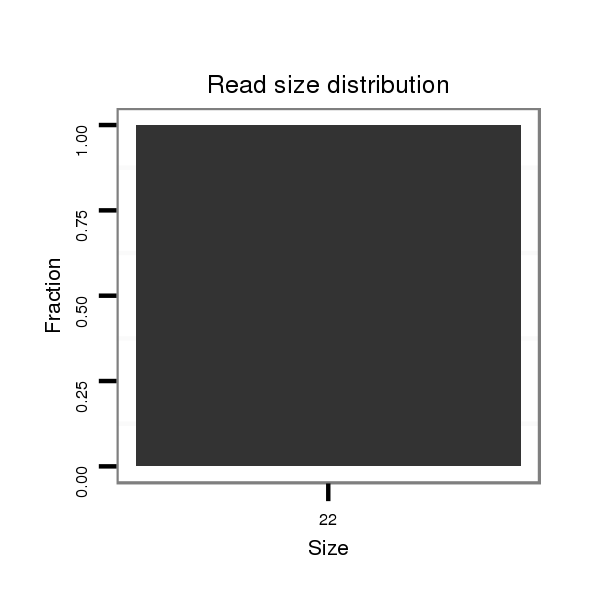
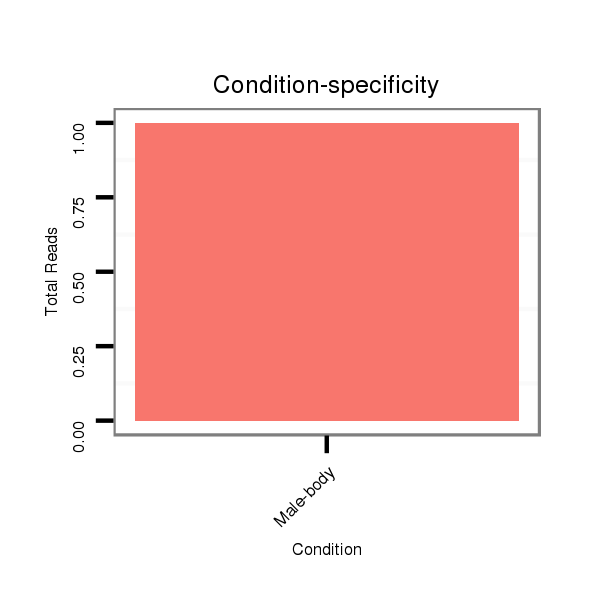
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060681 Argx9_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060670 9_testes_total |
V053 head |
M047 female body |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................AAGATCCGTACAGTTGACTCTG.................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................CAATACCAGCTGCATATCCAC.... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................CGTATGATGAGATGGAGA................................................................................................................................................................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................TCGTCCAGTTCATGTGCCTGTG.................................... | 22 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................TGCGAGGGTCGGTAGGTGAT........................................................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................TGTGGGTAGGGGAAGCTC.................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ........................................................................................................................AGCATGATCCGTACGGCT......................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................................................CGGGCGGCATGCTTAATAC................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TCACCTCACGCCCTAATGCGAACTACTCTACCTCTAGTGACGCTTACACCCATCCACTACCAGTTTACATTTGTCGGAATTCACCGACGACACGGCGAAACTATTCGATCGGGCGGTTCATCGTTCTAGGCATGTCAACTGAGACGTCGTCGATTAACACGGCGAATAGCACCGTGAAAAATAGTGTTCATAAACAACAAACAATTTATAAAACGTTAGCACGTCACGTACACGGACACCCGCCGTTGGAATTATGGTCGACGTATAGGTGTAGC
**************************************************....(((((((...........(((((......)))))..(((((((...(((((((......((......)).......(((((((((...(((...))).))))))))).))))))))))))))...)))))))(((((((((........)))))))))((((....)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|
| ....CTCACGCCCTAATGCGAACTA.......................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................CGTTGGGATTATGGTCGGCGT........... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................AAGACGTCGTCGATTAAC..................................................................................................................... | 18 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................TGCTAGGCAGGTCAAGTGAG.................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:8325218-8325492 + | dvi_18968 | AGTGGAGTGCGGGATTACGCTTGATGAGATGGAGATCACTGCGAATGTGGGTAGGTG--ATGGTCAAATGTAAACAGCCTTAA--GTGGCTGCTGTGCCGCTTT-GATAAGCTAGC---------------------------------CCGCCAAGTAGCAAGATCCGTACAGTTGACT--CTGCAGCAGCTAA-----------TTGTGCC------------------------GC-----TTATCGT----GGC-----ACTTTTTAT----------CACAAG----------------TATTTGTTGTTTGTTAAATATTTTGCA-ATCG------TGCAGTGCATGTGCCTGTGGGC------GGCAACC-TTAATACCAGCTGCATATCCACATC---------G |
| droMoj3 | scaffold_6680:11979639-11979953 + | TGG--ACTACTGGA--------AGTCCAATGGAAGTCAATCAGAATGTGGGTAGGTG--ATGACCACAA--------------------CTGAAGAGACTCTCT-GGAAAGCTGTCTCTTATGTACAAAATGCTATTGTGGGTTACTCCCGGACACGCAGCCTG--------AGTCAGCT--CTTCAGCAACTAA-----------ATGCGCA--GCTTAAGCGAACAAATAGTGTCACATTGCTTATCGTTTCG--CGTTGTTGTTTTTAT----------CGTAAT----------------TATTTGTTGTTTGTTTAACATTTCGCA-TAAT------CATAGTGCATGCGACCGTTGGC------GGTGGCC-TACATCCGAGCTGCACATCACCAAAATACACAGGC | |
| droGri2 | scaffold_15110:7353990-7354260 + | AGCTCA--------TTATTATTGTCGAGTTTTG---------GACGATGGGTAGGTGTAATGGTCAAATG-------CTCTAACTATGGCTGCTGAATGTCTTAAGATGAG-------------------------------------------------------ATGTACAGTTTACAAATTGC-GCAGCCAGCTGGGACTACTTTGTGCCTGACT-----G------------AACTGAGCTTATCGCTTTGTGT-----TCTTTTTAT----------CATAAGCAGCACAAATACATTGCATGTGTTATTTGCTTAATATTTCGCCAACGATAG---C--GACGCGAGCGCCGTTTGGA------GGTGACC-TC-GTAACAG--------CCACATC---------C | |
| droAna3 | scaffold_13337:12765811-12765984 - | CGAGGAGC--AAGAC---------------------C----------------------------------------------------------------------AGAGCGGTCTC----------------------------------------------------------------------------------------------------------GACAAACTGTGCCACTTGGCTAATCATCT-GCGCACTTTTA------TCTTGGCTAAGCTCCAT----------------CATGTGTTGTTTGCTCTATATTTCGCT-TTGCTAGCCCTG---CATCACGTTCCCTCCGAGGACTCAACGACCCCAAATGGCAGCCTGGGCTCCAC------------- | |
| droKik1 | scf7180000302686:72717-72849 + | AA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGTGCCGCCTGGCTAATCATTT-GCACTTTTTTA------TCTTGGCTA-------A----------------CATGTGTTGTTTGCTCTATATTTCCCG-ACGGCAGCCTCG---CGACGCAGCCCCTCGCC------GGACGCC---GATGACGACTCCGTCTCCAAGAC---------C | |
| droBia1 | scf7180000302193:3278100-3278170 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGTGTTGTTTGCTCTATATTTCGCT-ACGATAG------GACGCCGTAGCCCCTCGCC------AGCTGCG---------------ACCTCCCGGTC---------T | |
| droTak1 | scf7180000415868:479624-479718 + | CAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTA------TCTTGGCTAAGCTCCAT----------------TATGTGTTGTTTGCTCTATATTTCGCT-ACGATAG---CA---CGGCGTAGTCCCTCGCC------AGGCGGC---------------GTCTCCCAGTC---------T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/17/2015 at 03:53 AM