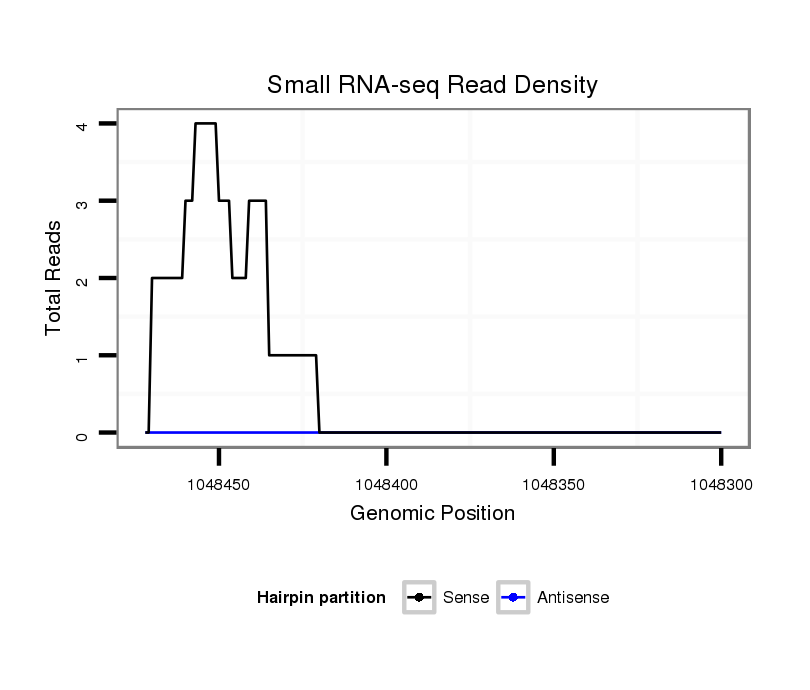
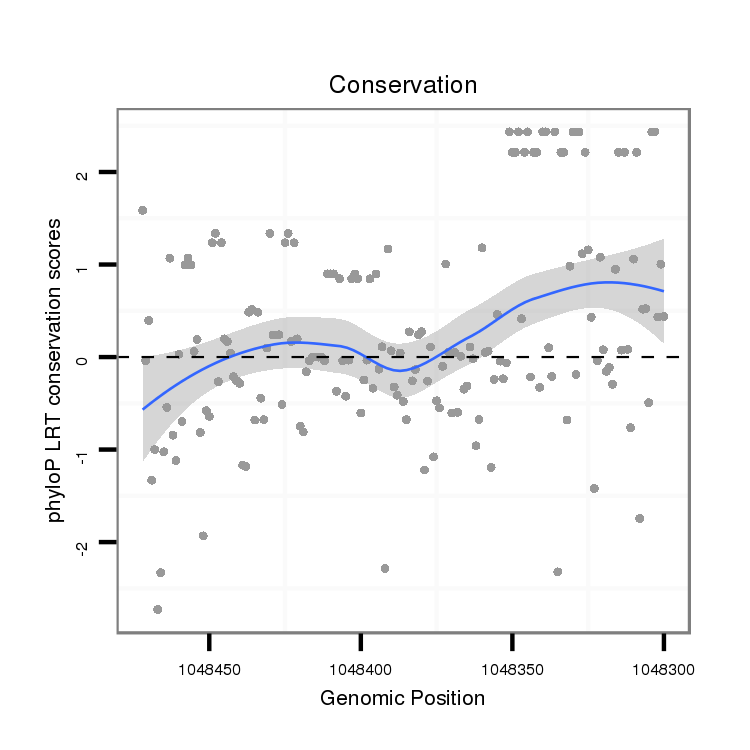
ID:dvi_1873 |
Coordinate:scaffold_12726:1048350-1048422 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_15569:1]; CDS [Dvir\GJ15248-cds]; CDS [Dvir\GJ15248-cds]; exon [dvir_GLEANR_15569:2]; intron [Dvir\GJ15248-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------################################################## ATGAGCCCGAGCTAGACGAGGACGATGACAATATAGATGCCGAATTCGAGGTACACAAAATATTAGCTGCACCCGGCTGACAAATTGCTTATTTAAACAAATTTCATTTGAATTGGATAACAGGTTGAGGCCATTGTGGGCCATAAGACCAAGCGCGGCGAATCGTATTTCCT **************************************************............(((((((....)))))))....(((.((((((((.(((((...)))))..)))))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
V053 head |
SRR060684 140x9_0-2h_embryos_total |
M047 female body |
M061 embryo |
SRR060662 9x160_0-2h_embryos_total |
SRR1106720 embryo_8-10h |
SRR1106722 embryo_10-12h |
SRR1106724 embryo_12-14h |
SRR060687 9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060656 9x160_ovaries_total |
SRR1106729 mixed whole adult body |
SRR060672 9x160_females_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060689 160x9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................GATGGATAACAGGTTGAG............................................ | 18 | 2 | 5 | 11.20 | 56 | 13 | 10 | 9 | 4 | 5 | 4 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| .................................................................................................................TGGATAACAGGTTGAGAT.......................................... | 18 | 2 | 2 | 5.50 | 11 | 0 | 3 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................ATGGATAACAGGTTGAGA........................................... | 18 | 2 | 5 | 2.60 | 13 | 2 | 1 | 2 | 2 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TGGATAACAGGTTGAGATT......................................... | 19 | 3 | 20 | 1.95 | 39 | 7 | 6 | 9 | 1 | 2 | 5 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 |
| ...............................TATAGATGCCGAATTCGAGGT......................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............ACGAGGACGATGACAATATAGA........................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............TAGACGAGGACGATGACAATATAGA........................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GAGCCCGAGCTAGACGAGGACGAT................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................TTCACTTGAATTGGATAACAG.................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AGCTGGATAACAGGTTGAG............................................ | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GAGCCCGAGCTAGACGAGGA....................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................ATGGATAACAGGTTGAGAT.......................................... | 19 | 3 | 20 | 0.95 | 19 | 8 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TGGATAACAGGTTGAGATCC........................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CAATGGATAACAGGTTGAG............................................ | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................ATTAGCTGCACCTGGTTCACAA.......................................................................................... | 22 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TATGGATAACAGGTTGAG............................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................ATTAGCTGCACCTGGCTCACAAT......................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GGATAACAGGTTGAGAT.......................................... | 17 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGATGGATAACAGGTTGAGG........................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TTAGCTGCACCTGGTTCACAA.......................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................GGTCGAGGCCATGGTGGG................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AGTAGGTTGAGGCCGTTGTG................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................ACATGGATAACAGGTTGAGA........................................... | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AGATTTCATTTGAATTGCAT....................................................... | 20 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................GACCAAGCGCGGGTAAACG........ | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ACCAAGCGCGGGTAAACGT....... | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TGATGGATAACAGGTTGAG............................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TACTCGGGCTCGATCTGCTCCTGCTACTGTTATATCTACGGCTTAAGCTCCATGTGTTTTATAATCGACGTGGGCCGACTGTTTAACGAATAAATTTGTTTAAAGTAAACTTAACCTATTGTCCAACTCCGGTAACACCCGGTATTCTGGTTCGCGCCGCTTAGCATAAAGGA
**************************************************............(((((((....)))))))....(((.((((((((.(((((...)))))..)))))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M028 head |
SRR1106727 larvae |
|---|---|---|---|---|---|---|---|---|
| ......................................CGTCCTAGGCTCCATGTG..................................................................................................................... | 18 | 3 | 20 | 0.50 | 10 | 10 | 0 | 0 |
| .....................................ACGGCTTAAGCTCAATGTTC.................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ...................................................................................................................CTCTTATCCAACTCCGGCA....................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:1048300-1048472 - | dvi_1873 | ATGAGCC---------------CGAGCTAGACGAGGACGATGACA---ATATAGATG---------------------------------C---CGAATTCGAGGTACACAAAATATTAGCTGCACCCGGCTG---------------------------ACAAATTGCT-----TATTTAAACAAATTTCATT----------------TGAATTGGATAACAGGTTGAGGCCATTGTGGGCCATAAGACCAAGCGCGGCGAATCGTATTTCCT |
| droMoj3 | scaffold_6473:9114338-9114515 + | dmo_717 | ATGACAA---------------TAATGATGACAATGACGATGACA---ACGCC---------------AACGCCGATGATGATGTTGACAG---GGAATTCGAGGTT--------------TGCACACGGTTAAAATAC---------------------------TTGC-----CATAAG------GCGAAC---------TATTATATTCGTTTGCATTACAGGTGGAGGCGATTGTAGGTCACAAGTCGAAGAACGGCGAGTCGTTTTTCCT |
| droGri2 | scaffold_14853:10002475-10002633 - | ATGACAA---------------GGAGAAGGACGAGACGGACGACGAAAGTGGGGAGG---------------------------------C---AGAATTTGAGGTAAGC----TATTTGCA---------------------------TTTGCAGGAGAATAAGACGATTTTATTATTGAATC----------------------------------TATCTAGGTGGAGGCCATCGTGGGTCACAAGACCAAGAACGGAGAATCATTTTTCCT | |
| droWil2 | scf2_1100000004909:7203625-7203787 + | ATGTAGCCGATGGGGGAGAAG------------AACCGGAAGCAGA------------------------------------AGATGAAAA---GGAATTTGAGGTTTGT----CATTAGTT---------------------------TTCGAA-----ACAAATCAGT-----TTTTTAAGTCAAGTGCTCTCTTT--------------------TTAAAAGGTGGAAGCCATTGTTGGTCATAAGACGATAAAATGCGTCTCGTACTTCTT | |
| dp5 | XL_group1e:4445150-4445387 + | dps_2682 | AAGATTCAGATTCGGATGATGACTACGAGGACGCTATCGATGACG------------AATCCTTAGCTAAGTCCGATGACGATTTTGACACAAAGCAGCAAGATGCC--------------GACACAGAGTTGGAGTACGAAGTGAGTGATAGAG-----TG-GGACAGT-----CGCTCTGCGTTAGAGTTTGCTTTAACCTAAAAAAATCTGTAAATTTCTAGGTGGACGCCATTCTTGGCCACAGGACGGTGCGTGGCGCCTCCTATTTTCT |
| droPer2 | scaffold_17:1387260-1387483 + | ATGGTGA---------------CTACGAGGACGCTGTCGATGACG------------AATCCTTAGCTAAGTCCGATGACGATTTTGACACAAAGCAGCAAGATGTC--------------GACACAGAGTTGGAGTACGAAGTAAGTGATAGAG-----TAAAGACAGT-----CGCTCTGCGTTAGAGTTTGCTTTAACCTAAAAAAATCTGTAAATTTCTAGGTGGACGCCATTATTGGCCACAAGAAGGTGCGCGGCGCCTCCTATTTTCT | |
| droAna3 | scaffold_13248:1824927-1824978 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGTGGAAGCCATTATTGGGCACAAGACTGTGCGTGGCGTTTCGCACTTCCT | |
| droBip1 | scf7180000395544:37352-37404 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTGGAAGCCATTGTGGGACACAAGACTGTGCGCGGCGTCTCCTATTTCCA | |
| droKik1 | scf7180000302592:1540188-1540267 + | AAATTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAATTAAACCATTATATTCCAGGTGGAGGCAATTGTCGGTCACAAGACTTTGCGCGGCGCCTCATACTTTCT | |
| droEle1 | scf7180000490996:231511-231563 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTAGAAGCGATCATGGGCCACAAGATTATGCGCGGCGCATCGTATTTCCT | |
| droRho1 | scf7180000779333:759428-759479 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGTGGAGGCGATCATAGGCCACAAGACAATGCGCGGCGCATCGTACTTCCT | |
| droBia1 | scf7180000301760:3547210-3547262 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTGGAGGCCATCGTGGGCCAGAAGACGGTGAAGGGCGCCACCTATTTCCA | |
| droTak1 | scf7180000415210:498590-498642 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTGGAGGCCATTGTGGGCCAGAAGACGGTTAAGGGAGCCACCTACTTCCA | |
| droEug1 | scf7180000409271:601223-601274 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGTGGAGGCGATCGTCGGCAAAAAGACGGTTAAGGGTGCTACACACTTCCT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 06/11/2015 at 05:58 PM