
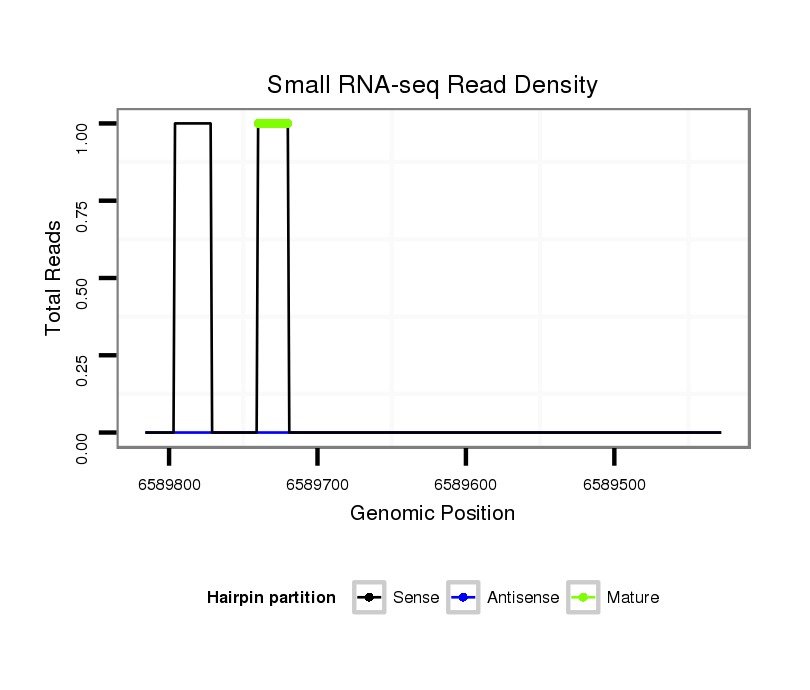
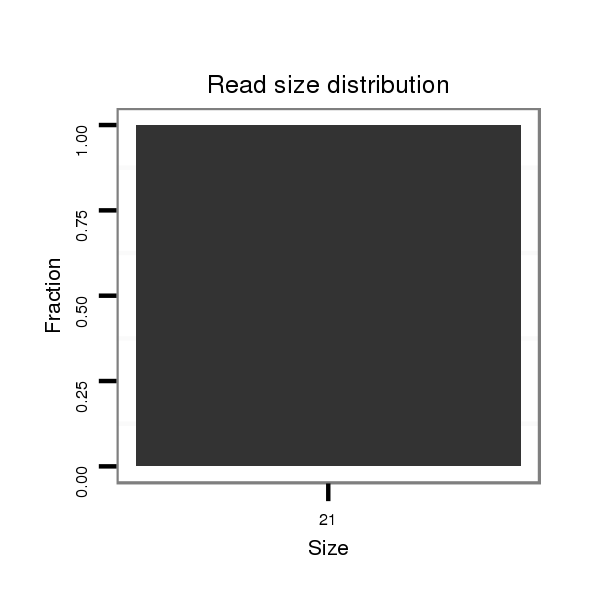
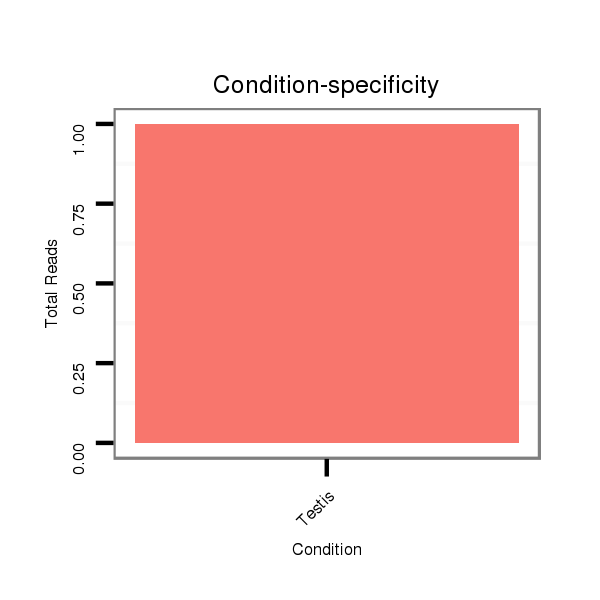
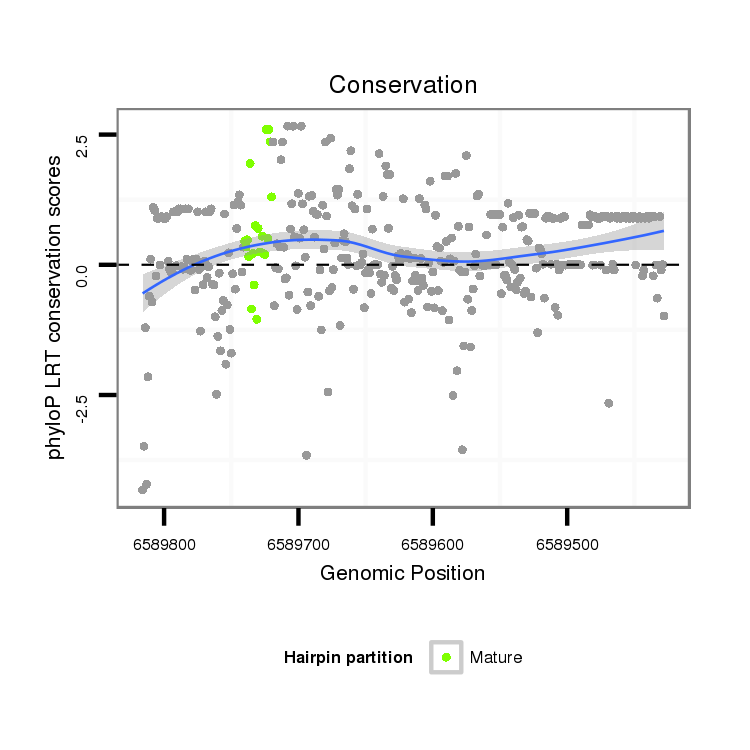
ID:dvi_18700 |
Coordinate:scaffold_13049:6589478-6589766 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -58.5 | -58.3 | -58.2 |
 |
 |
 |
CDS [Dvir\GJ12291-cds]; exon [dvir_GLEANR_12269:9]; CDS [Dvir\GJ12291-cds]; exon [dvir_GLEANR_12269:10]; intron [Dvir\GJ12291-in]
| Name | Class | Family | Strand |
| (CAGT)n | Simple_repeat | Simple_repeat | + |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACGTGGAGACGCGTTCCCTTTATCGCGATGACTCTCCAGGCGTGGTACAGGTGTGGTATATCTATATAACTGACTGACTGATTGAGTGGCTGACTGACAGAGTGACAGATAGATTGACTGATGAATTCGCTAAGTGATTGAGTGACTGATGCACTGACTAAGCAGTCACGTTCTGACCAACTGGCAAGTAATTGACTAGCTCATGGACTAACTGATGTACAAGTGACTAACTGATTGAGAAAGTAATTGAACTTTAGATAGATGTTACCGCGAACAGTCTAACATTTGAATAAAAGCCCGCTAACGCCGGACTTTAACTCGCTTCACTTTCGATTTTAGGGTAAAATTCAGATGTGGATAGAGCTGTACGAGCAGAACATGTTCATTCC **************************************************....(((..((((((....(((....(((.....(((.....))).....)))..)))))))))..)))..((((.(((..((((((.((((((((((.(((..((((..((((((((.(((((..((....))..)).)))))))).)))......(((.((((.((.(((...(((..((.....))..)))..))).)).)))).)))..))))..)))..))))).............((((((.((....)).)).)))).)))))..)))))).))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M028 head |
M027 male body |
V053 head |
M061 embryo |
SRR060684 140x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
M047 female body |
V047 embryo |
V116 male body |
SRR060663 160_0-2h_embryos_total |
SRR060670 9_testes_total |
SRR060681 Argx9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060655 9x160_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060674 9x140_ovaries_total |
SRR060671 9x160_males_carcasses_total |
SRR060683 160_testes_total |
SRR060658 140_ovaries_total |
SRR060654 160x9_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060680 9xArg_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................................................................................ACTTTCGATGGTAGGATA.............................................. | 18 | 3 | 20 | 8.70 | 174 | 0 | 40 | 0 | 1 | 25 | 26 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 16 | 0 | 8 | 1 | 7 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................CTGACTGTGAGAGTGACAGCT....................................................................................................................................................................................................................................................................................... | 21 | 3 | 9 | 5.67 | 51 | 28 | 2 | 4 | 12 | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGACTGTGAGAGTGACA.......................................................................................................................................................................................................................................................................................... | 18 | 2 | 12 | 2.92 | 35 | 15 | 2 | 6 | 0 | 0 | 0 | 0 | 9 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGACTGTGAGAGTGACAG......................................................................................................................................................................................................................................................................................... | 19 | 2 | 9 | 2.11 | 19 | 10 | 2 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GCCTGACTGTGAGAGTGACAGAT....................................................................................................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACTGATTGAGTGGCTGACTGA.................................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TATCGCGATGACTCTCCAGGCGTGG........................................................................................................................................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GCCTGACTGCGAGAGTGACAG......................................................................................................................................................................................................................................................................................... | 21 | 3 | 11 | 0.82 | 9 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GCCTGACTGCGAGAGTGACA.......................................................................................................................................................................................................................................................................................... | 20 | 3 | 17 | 0.76 | 13 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GATGAACTCGCAAGGTGATT.......................................................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CTCCAGAGGTGGTAGAGGT................................................................................................................................................................................................................................................................................................................................................. | 19 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................GGCTGACTGTGAGAGTGACAGCT....................................................................................................................................................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TTGATCTTTAGAAAGATGCTAC......................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................ACTTTCGATGGTAGGGTA.............................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGACTGCGAGAGTGACAGCT....................................................................................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGACTGCGAGAGTGACA.......................................................................................................................................................................................................................................................................................... | 18 | 2 | 12 | 0.25 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................ACTTTCGATGTTAGGATA.............................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTGACTGTCAGAGTGACA.......................................................................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGACTGCGAGAGTGACAG......................................................................................................................................................................................................................................................................................... | 19 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................TTGAATTGGAGCCCGCTAAC.................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACCGATTGAGTGGCTTAGTGA.................................................................................................................................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................GATTGAGTGGCTTAGTGA.................................................................................................................................................................................................................................................................................................... | 18 | 2 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................CACGTAATTGACTAGCTTTTG........................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TGACTGCGAGAGTGACAGCT....................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GATGAACTCGCAAGGTGAT........................................................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TTCTGACCAATAGGCACGT........................................................................................................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................CACTTTCGATGGTAGGATA.............................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................CGAACGGTTTAACATTCGA.................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................AACTTTCGATGGTAGGGTA.............................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................ACTTTCGATGGTAGGATAA............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................GCGATAGTAATTGAACTTA...................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTGACTGCGAGAGTGACA.......................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................AACTGGCAAGGAATTGGCGA............................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TGACTGCGAGAGTGACAG......................................................................................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGCACCTCTGCGCAAGGGAAATAGCGCTACTGAGAGGTCCGCACCATGTCCACACCATATAGATATATTGACTGACTGACTAACTCACCGACTGACTGTCTCACTGTCTATCTAACTGACTACTTAAGCGATTCACTAACTCACTGACTACGTGACTGATTCGTCAGTGCAAGACTGGTTGACCGTTCATTAACTGATCGAGTACCTGATTGACTACATGTTCACTGATTGACTAACTCTTTCATTAACTTGAAATCTATCTACAATGGCGCTTGTCAGATTGTAAACTTATTTTCGGGCGATTGCGGCCTGAAATTGAGCGAAGTGAAAGCTAAAATCCCATTTTAAGTCTACACCTATCTCGACATGCTCGTCTTGTACAAGTAAGG
**************************************************....(((..((((((....(((....(((.....(((.....))).....)))..)))))))))..)))..((((.(((..((((((.((((((((((.(((..((((..((((((((.(((((..((....))..)).)))))))).)))......(((.((((.((.(((...(((..((.....))..)))..))).)).)))).)))..))))..)))..))))).............((((((.((....)).)).)))).)))))..)))))).))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M027 male body |
SRR060662 9x160_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
SRR060680 9xArg_testes_total |
V116 male body |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060665 9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................................................TGGGGCTTGCCAGATTGTGA....................................................................................................... | 20 | 3 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................CGAAGTGAAAGCTAAAATCGT................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GCACGGGATATAGCGCAACT...................................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TGAGGGGTCCGCCCCATG..................................................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................AAGTGGAAGCTTAAATCACA............................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TCGTCAGTGCGAGACTGC................................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................GGGCGAGTGCGGGGTGAAATT........................................................................ | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................TTCGGGCGCTAGCAGCCTG............................................................................. | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................CTTTTTTTCGGGCAAGTGCG.................................................................................. | 20 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ............................................................AGATATATTGAATGACGGAGT.................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................GCTTGTCTGATTTTAAACTTG.................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................GGGCGACACCGGCCTGAAA.......................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................GTCCCATTTTAAGGCTACT.................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................AGAAATGGCGCGAAGTGAAA........................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................ACATGTTCGATGATGGACT........................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................GGGTGATACCGGCCTGAA........................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:6589428-6589816 - | dvi_18700 | ACGTGGAGACGCGTTCCCTTTATCGCGATGACTCTCCAGGCGTGGTACAGGTGTGGTAT---A--TCT-------------ATATAACTG-----------------------------------------------------------------------------------------------ACTGACTGATT----GAGTGGCTGACTGACAGAGTGACAG----ATAGATTGACTGATG--------AATTCGCTA-------------------------------------------------------------------------------------------A-------------------------GTGATTG---------------------------------AGTG-------ACT--------------GATGCACT------------GACTAAGCAGTCACGTTCTGACCAACTGGCAA-G-TAATTGACTAGCTCATGGACT----A-------------------------------------ACTGAT-GTA-CAAGTGACTAACTGATTGAGAAAGTAATT-----GAACTTTAGATAGATGTTACCGCGAAC--AGTCTAACATTTGAATAAAAGCC----------------------------------------------------------------------------CGCTAACGCCGGACTTTAACTCGCTTCACTTTCGATTTTAGGGTAAAATTCAGATGTGGATAGAGCTGTACGAGCAGAACATGTTCATTCC |
| droMoj3 | scaffold_6491:100319-100520 - | TGACT--------------------------------------------------------------------------------GACTG-----------------------------------------------------------------------------------------------ACA------------GAGCGACGAAATGACCGAGTGACAG----ATTGACAGAGTGACA--------GAGTCACTG-----AA---------------------------------------------------------------------------------------------------------------CAACAGGACGACT---------------------GACTGACAGAGCGG-----CG--------AAAT--------------------GA-------------------CAGAGTGACAGAG-CCACTGAACAACAATACAACT----G----------------ACTGACAG-----------------AA-----CAACGAAATGACAGAGTGACAGAGCCACT-----GAA--------------------GAAC--AGTACA---ACTGACTAAAAGACCG--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15074:7013758-7013931 + | TAACT--------------------------------------------------------------------------------GACTG-----------------------------------------------------------------------------------------------ACTAACT--------GACTGACTAACTGACTG----ACTA----ACTGACTGACTAACT--------GACTGACTA-----ACTGA----C-------------------------------------------------------------------------------------------------------TGACTAACTGACT---------------------G----ACTAACTGA-----CT--------AACT--------------------GA-------------------CTAACTGACTGAC-T------------------------A----ACTGACTGACTGACT-------------ACCTGACCAAC---------TGACTGACTGACTGGCCAAGTGACTAACTTGA------A--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:11030981-11031090 + | ATGGA---------------------------------------------------------------------------------ATTG-----------------------------------------------------------------------------------------------ACTGACTGAAT----GACTAACTGACTGACAG------------ACTAACGGACTGACT--------GACTG--------------------------------------------------------------------------------------------------------------------------------ACTAACT---------------------G---------------------------------ATTGGGTGGGTGGTTGTGCGGGTGA--AGCAATTGT----CTGAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------CA | |
| dp5 | XL_group1e:5228760-5228910 + | TGAT------------------------------------------------------------------------------------TG-----------------------------------------------------------------------------------------------ACT------------CAATGACTGACTGACTGAAGGATAT----ATTGACTAACTGATG--------GTTGGA----------------------------------------------------------------------------------CTTCATTCGGTAAATGAATGAAT--GACGGTTGGTTGGTT---GGACTCACTGACTGACGGATTGACG-----G----AT-------TGACT--------------GATTGACTGACT--------GA-------------------CTGAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------AGACGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_15:638317-638451 - | CTGAA-----------------------------------------------------------------------------------TG-----------------------------------------------------------------------------------------------ACTGACTGACT----GACTGACTGACTGACTG------------ACTGACTGACTGACC--------GATTGACTG-----ACTGTTTGAT-------------------------------------------------------------------------------------------------------TCACAGACTCACTGAGT--------GAATGACTG-----A-------------A--------TGAGT--------------------GA-------------------CTGAGTGACT-----------------------------G----ACTTGCTA----ACC-----A---------------------------------------------AA----------------------------------------------------------TAACCG--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13117:2727889-2728008 - | ATATCT--------------------------------------------------------------------------------ATAG---------------------------------------------------------------ATA------------------TGTATATTG--ATTGACTGATT----GACTGTCTG--------------------ACTGACTGACTGTCT--------GATTGACTG-----AC---------------------------------------------------------------------------------------------------------------TGACTGGCTGACT---------------------GACTGACTGATCGC-----TG--------A------------TCT--------GACTGA--TGACATGA-----------------------------------------------------------------------------------------------------------------------TCT-------------G--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000394714:86149-86252 - | TGAAT--------------------------------------------------------------------------------GATTG-----------------------------------------------------------------------------------------------ACT------------AATTGAATGATTGACTAACTGAATG----ATTGACTAACTGATT--------GATTGATTG----------------------------------------------------------------------------------------------------------------------------ACTAACTGACT-----------------A----AC-------TGACT--------------GACTGACTGACT--------GA-------------------CTGAC----C-----------------------------G----A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C | |
| droKik1 | scf7180000302577:1524033-1524183 - | ACTA----------------------------------------------------------------------------------ACTG-----------------------------------------------------------------------------------------------ACTGAATGAAT----GACTGACTGACTGGCGAACTGACTG----ACTGAATAACTGACG--------AACTGACGA-------------------------------------------------------------------------------------------A-------------------------CTGACGAACT----------------GACGAACTG-----A-------------C--------TGACTCGCTGGATGACT-------------------------------AAC-----------------------------------------TCACCA----AC----------------------------------------------TGAGTGAGTACTT-----AAAGCAATGGCAAACATTA----------------------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453872:503544-503653 - | CCAAT-----------------------------------------------------------------------------------TG-----------------------------------------------------------------------------------------------CCTGATTGACT----CACTGGCTGACTGACTGACTGACTG----ATTGACTAATTGACT--------AAATGACTG---------A----T-------------------------------------------------------------------------------------------------------TGACTAACTGGCT---------------------G----AT-------TGACT--------------TGCTGACTGACT-------------------------------GAC----T-------------------------------------GA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CG | |
| droEle1 | scf7180000491006:1189547-1189727 - | ACAGCT---------------------------------------------------------------------------AAGTAATTG-----------------------------------------------------------------------------------------------ACT------------AACTGACTAACTGACTAACTGACTA----ACTGACTAACTGTCT--------AACTGACTA-----AC---------------------------------------------------------------------------------------------------------------TGACTAACTGACT---------------------A----AC-------TGACT--------------AACTGACTATCT--------GA-------------------CTAACTGACTAGTTAGTCACAATGTGCGACTTAGTGACTA----TTTGGCTA----ACT----------------------------------GACTTACTGAATATCTCACTATTA-------------G--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779914:688598-688816 + | CTAACT-------------------------------------------------------------------------------GACTG-----------------------------------------------------------------------------------------------ACTGACT--------GATTGACTGACTGACTGACTGACTG----ACTGACTGACTGACT--------GACTGACTG---------A----C-------------------------------------------------------------------------------------------------------TGACTGACTGACT---------------------G----AC-------TGACT--------------GACTGACTGACT--------GA-------------------CTGACTGACTG---------------------------------ACTGACTG----ACT-----GACTCACTGACTCACTGACTGAA-TGACTAACTCGCCAACTGAGTGAGTACTT-----AAAGCAATGGCAAACATTAGCATAAAGCTGGGCTATCA-----------------------------------------------------------------------------------------------------------------------------T-----------------------------------------------------TT | |
| droBia1 | scf7180000301760:732261-732410 - | GACTG---------------------------------------------------------------------------------ATGG-----------------------------------------------------------------------------------------------ACTGACT------------GATGGACTGACTGGGTGACAGACTGACTGACTAGCTGACC--------AATTGGCTG-------------------------------------------------------------------------------------------A-------------------------CTGACTG---------------------------------AGAG----G-----CCAACAGACAGACT--------------------GA-------------------CTGGCTGACTGGC---------------------T----GACTGATTGACTG----ACT----------------------------------GACTGACTGATTGATTGACTGACT-------------G--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000410393:20894-21123 - | CT-----------------TCATCGCGATAGCTTTAAAACTGAGAGACTAGTTCGCATA---GAAACG-----GACAGACTGACAGACTG-----------------------------------------------------------------------------------------------ACA------------GACTGACAGACTGACAGACTGACAG----ACTGACAGACTGACAGACTGACAGACTGAC---------AGA----C-------------------------------------------------------------------------------------------------------TGACA-----------------------------GACTGACAGACTGA-----CA--------GACT--------------------GA-------------------CAGACTGACAGAC-TGACAGACTGACAGACTGACA----GAC----------------TGACAG-----------------AC-----TGACAGACTGACAGACTGACAGACTGACA-------------G--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000408940:234-503 + | ACTGACAGAC--------------------------------------T------------------------GACAGACTGACAGACTG-----------------------------------------------------------------------------------------------ACA------------GACTGACAGACTGACAGACTGACAG----ACTGACAGACTGACAGACTGACAGACTGAC---------AGA----C-------------------------------------------------------------------------------------------------------TGACA-----------------------------GACTGACAGACTGA-----CA--------GACT--------------------GA-------------------CAGACTGACAGAC-TGACAGACTGACAGACTGACA----GAC----------------TGACAG-----------------AC-----TGACAGACTGACAGACTGACAGACTGACA-------------G---------------------------------ACTAACAGACTGACAGACTGACAGACTGACAGACTGACAGACCTTGTTTAACTTTTTTCTGTTTATATC----------------------------------------------TTTCGAT-----------------------------------------------------TT | |
| dm3 | chr3L:8900297-8900673 - | ATGTGGAAACCCGCTCCCTTTGCCGCGATGATTTTCCAAATGTCGAACAGGTAAGTTAG---GCAACGAAGTG------------------------TAGGTGCAAACCTATCCCAATGCAATGACTCACTCACATAGTTAGAATATCTAAAAATAGCATGGCCAAATTAAAGCTATACATATAT--GGCCAAAAAGTGTCTAGA--------------------------------------------------------CAGTCTCTATCAAT-A---TTCAACTAATCTGTGAGAAATA------TGT-TGATA-AATGTTGATAAAACATATTACGTTTTATTTTCGATATAAAAAAGTATTATTTTTATTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------TATA----------------------------------------------------------------------------------TATTTATAATAAACCAAACTACTAATGCAACCCATTG------------------TCCAGGGCAAACTACAACTGTGGATAGAGCTGTTCGAGGCGAACATCTATGTGCC | |
| droSim2 | x:14689665-14689851 + | GTGGGGGC-----------------------------------------------------------T-------------GACTAACTG-----------------------------------------------------------------------------------------------ACTAACT------------GACAGACTGACTAGCCGACTT----AGCGACTAACTGACC--------AACTGACTA-----ACTGACAAAC-TGGCTT---------------------------------------------------------------------A-------------------------CTGACTA--------AGT--------GACTAACTG-----A-------------C--------TTACT--------------------GA-------------------CTAAGTGACT-----------------------------A----ACTGACTA----ACT-----G----ACTAACTGACAGAC-ACACTAGCTGACTAGCCGACTTAGCGACTAACT-------------G--------------------------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_0:1170821-1171205 - | ATGTGGAAACCCGCTCCCTCTGCCGCGATGATTTTCCAAATGTTGAACAGGTAATAAAA---ACAACGAAGTG--TAGTGT--------------------AGCAAACTTATCCCAATGTAATGACTCACTT--TTAGTTAAAATTTCTACAAATAGCATTGCCAAATTAAAGCTATACATTTAC--GACCAAAAACAGTCTAGA--------------------------------------------------------GAGCCGTTATCAAT-ATTGTTTACCTAATCTGTGACAAATTGCATTATGAGTAATA-ATCATTGATAAAACATATTTCGTTTTATTTTAGATATAAAAAAGTAATATTTTCATTAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------TATA------------------------------------------------------------------------------------TTTATAATAAACCAATCTACTAATGCAACCCATTG------------------TCCAGGGCAAACTCCAACTGTGGATAGAGCTGTTCGAGGCGAACATCTATGTGCC | |
| droYak3 | 3L:3013562-3013892 + | ATGTGGAAACCCGCTCCCTTTGCCGAGATGATTTTCCAAATGTCGAACAGGTAATA--GGAAACA------------------------------ATTACGCGCTAACCTATCCCAATGAAATGACTCATTG--ATGGTAAAAATATCCAAAAATA------------------TGTACATCTAT--TGCCATAAAAAATCATG--------------------------------------------------------------------AAA-ATGAATCAGCTTATCTGTGACAAATGGCATTTTAGCTAGCA-ATCATAGATAACA-TTATGTAGTTTAATTTGAGATATAAATTTGT--TTTTTTTGGTTAAGT---AAACT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTAATGCAAACCATTG------------------TCCAGGGCAAACTGCAACTGTGGATAGAGCTGTTCGAGGCGAACATCTATGTGCC | |
| droEre2 | scaffold_4784:5143106-5143441 + | ATGTGGAAACCCGCTCCCTTTGCCGAGATGATTTTCCCAATGTCGAGCAGGTAATTTAA---ACACCAAAAGG--C--TTTTATGAGTTAACCAAATTACGCGCTTACCTATCCCAATGAAGTGACTCATTT--GTAGTAAAAATATC--------------------------TGTACATGTAT--TGCCATAAGGAGTCTAGA------------------------------------------------------------------CAAA-ATTAACCACCTAATCAGTGACAAACAGCATTACAAATAGCGCATCATTGATAACA---------------------------------------------G----------------------------------------------------------------------------------------------A--TAG-------------------------------------------------------------------------------------------AT-----TT--------ATATAAAATTAGAGTTTT-----------------------------------------------------------TACATA------------------------------------------------------------------TTGTGCTAATGCCACCCATTG------------------TCCAGGGCAAACTCCAACTGTGGATAGAGCTGTTCGAGGCGAACATCTACGTGCC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:49 PM