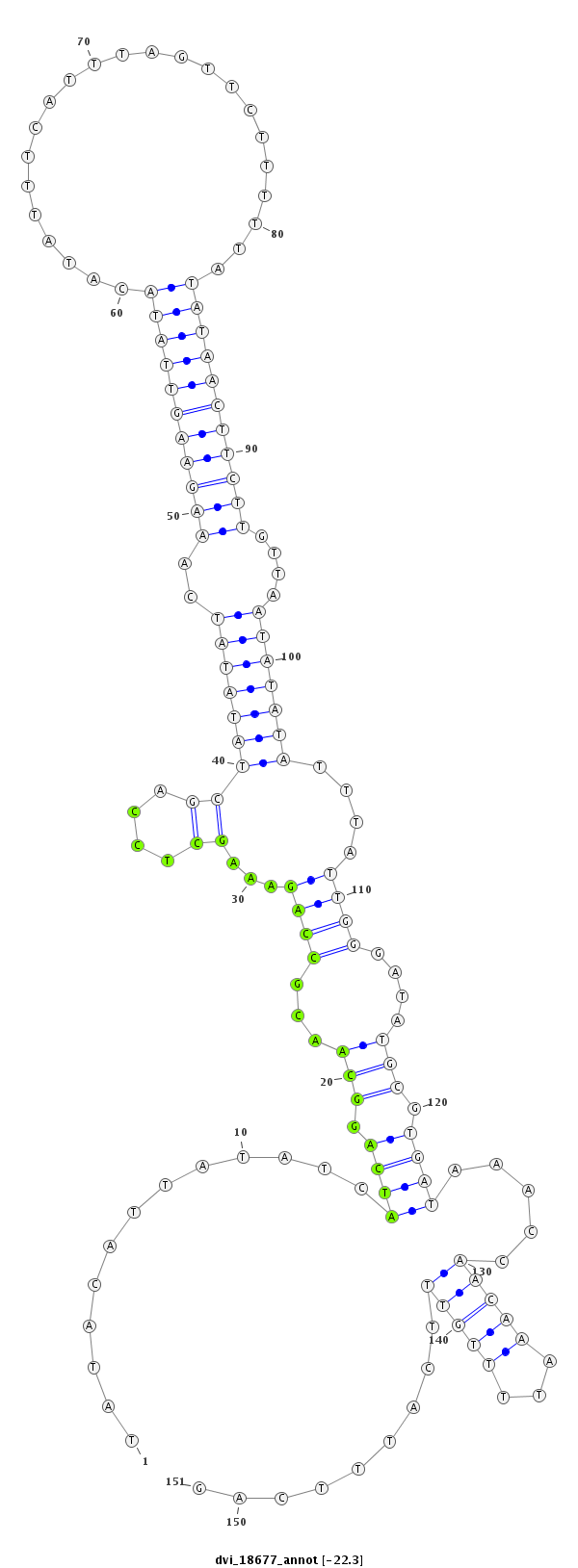
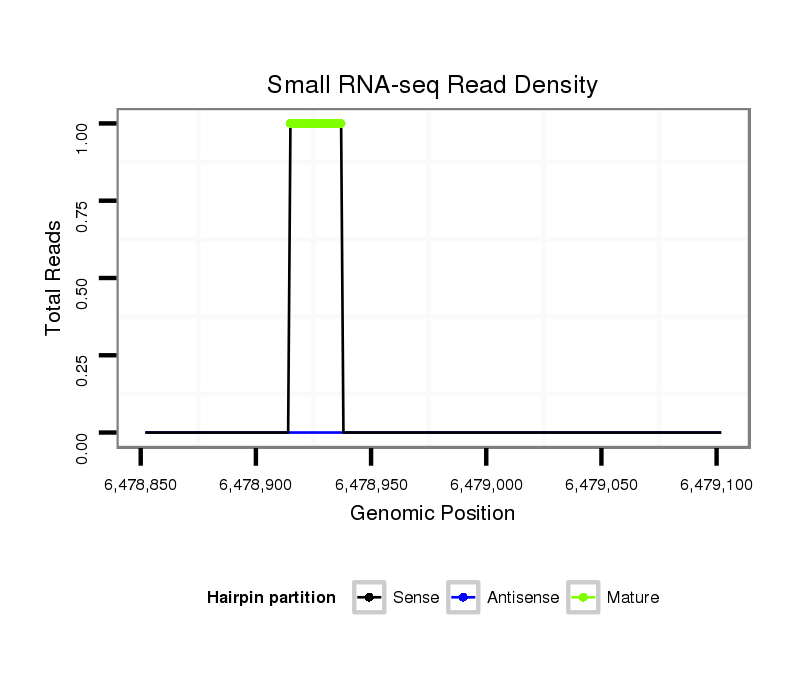
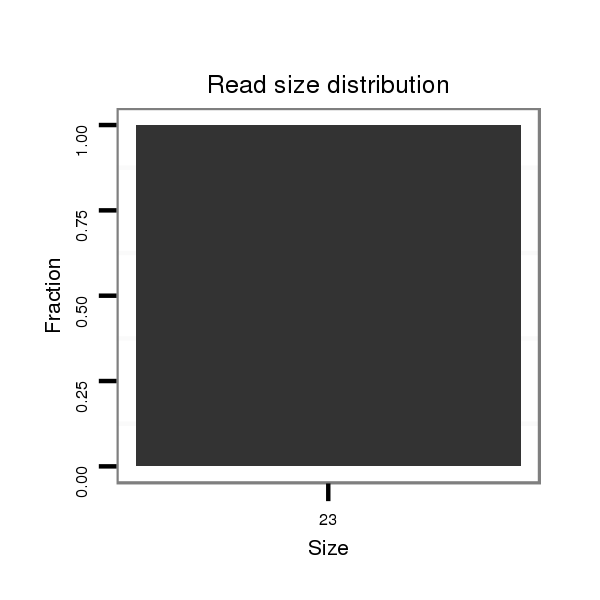
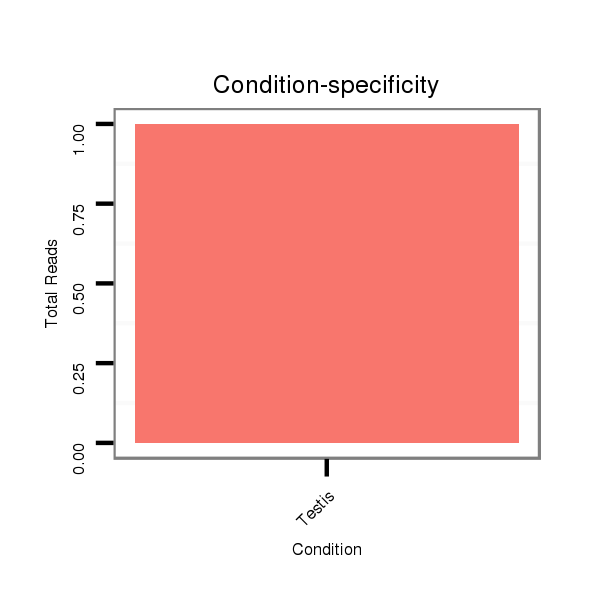
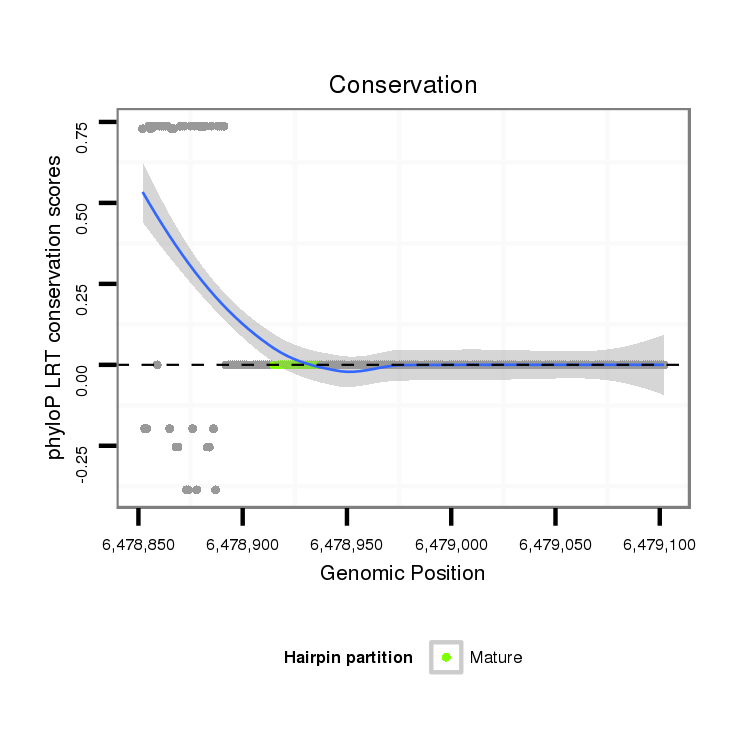
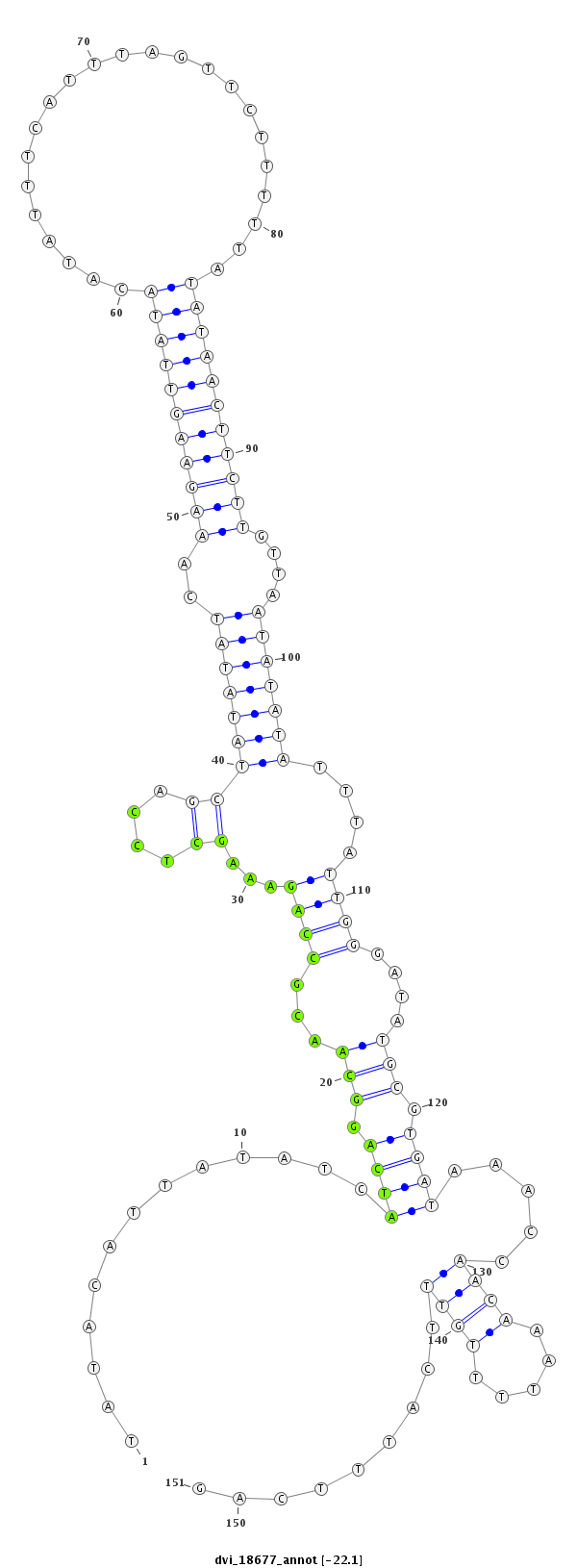
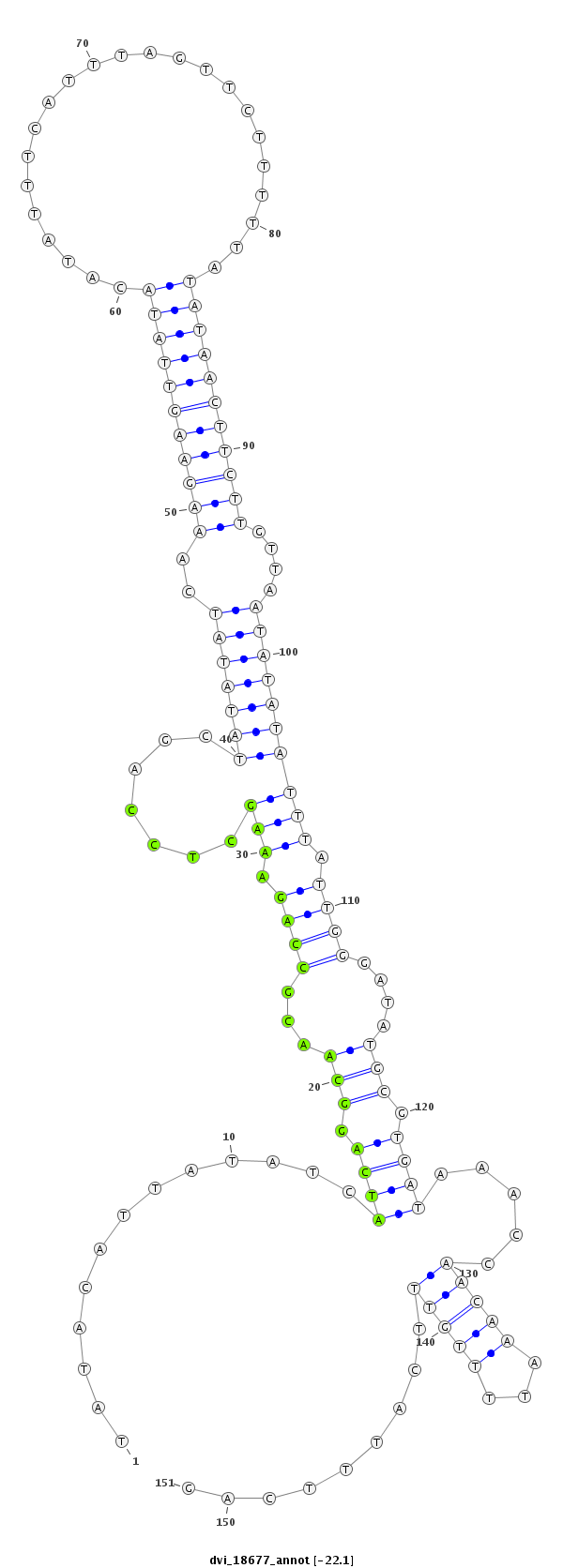
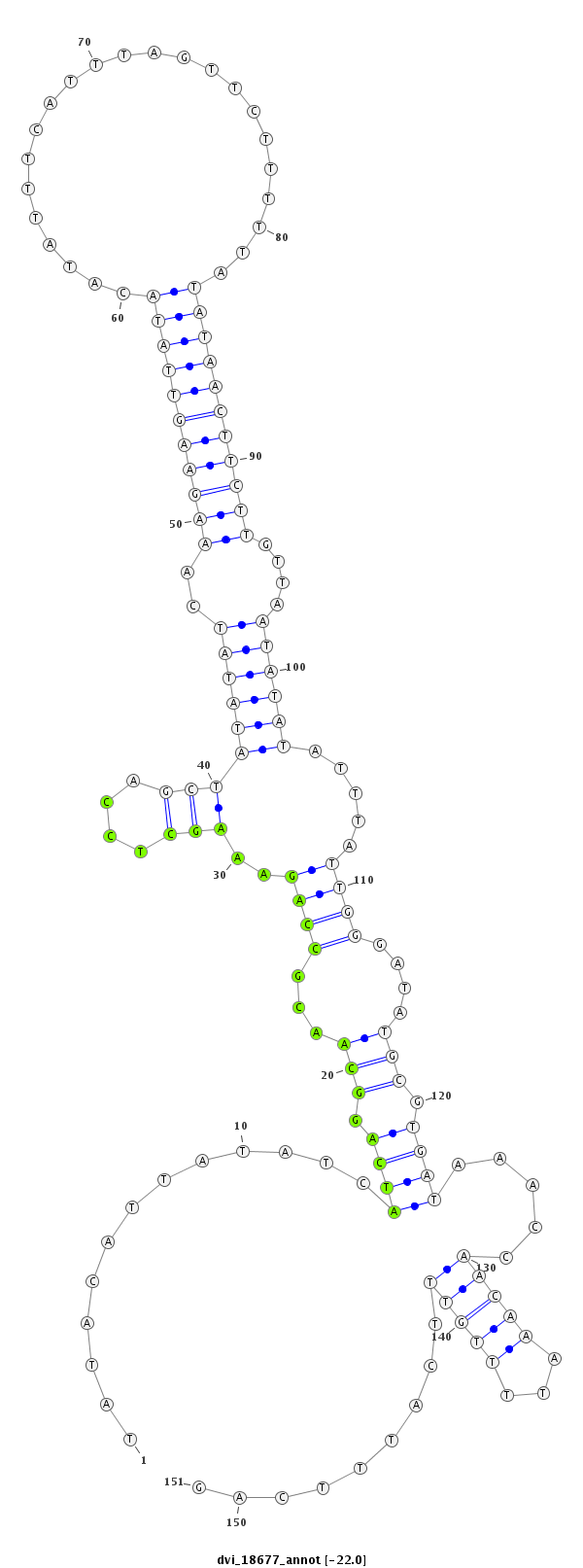
ID:dvi_18677 |
Coordinate:scaffold_13049:6478902-6479052 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.1 | -22.1 | -22.0 |
 |
 |
 |
CDS [Dvir\GJ13076-cds]; exon [dvir_GLEANR_12996:3]; intron [Dvir\GJ13076-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTTACTAATTTTTGGCTTAAATCTCATATTATTAGATAATATTTTATTTATATACATTATATCATCAGGCAACGCCAGAAAGCTCCAGCTATATATCAAAGAAGTTATACATATTTCATTTAGTTCTTTTTATATAACTTCTTGTTAATATATATTTATTGGGATATGCGTGATAAACCAACAAATTTTGTTTCATTTCAGATTTCCACGTATTTTCCGCAGTCGCAGGTGGAACTGCCCAAAACCTCGTC **************************************************.............((((.(((...((((...((....))(((((((..(((((((((((.......................)))))))))))....)))))))....))))....))).)))).....(((((...))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
M028 head |
SRR060663 160_0-2h_embryos_total |
V116 male body |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................AGACACCGGTGGAACTGCCC........... | 20 | 3 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................ATCAGGCAACGCCAGAAAGCTCC..................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................AACAGCTTTTCTTTCATTTCAGAT................................................ | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................GGGCGATGCGGGATAAACC........................................................................ | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................CACAAAGGTCCAGCTAGATAT........................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................GTCGAAAGTGGAACAGCCC........... | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TTTCGAAGTCGCAGGTGGCA................. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................GGCGATGCGGGATAAACC........................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
GAATGATTAAAAACCGAATTTAGAGTATAATAATCTATTATAAAATAAATATATGTAATATAGTAGTCCGTTGCGGTCTTTCGAGGTCGATATATAGTTTCTTCAATATGTATAAAGTAAATCAAGAAAAATATATTGAAGAACAATTATATATAAATAACCCTATACGCACTATTTGGTTGTTTAAAACAAAGTAAAGTCTAAAGGTGCATAAAAGGCGTCAGCGTCCACCTTGACGGGTTTTGGAGCAG
**************************************************.............((((.(((...((((...((....))(((((((..(((((((((((.......................)))))))))))....)))))))....))))....))).)))).....(((((...))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
V053 head |
SRR060678 9x140_testes_total |
SRR060683 160_testes_total |
SRR060667 160_females_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060673 9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060666 160_males_carcasses_total |
M027 male body |
SRR060661 160x9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................AATATATTGAACAACAATTA...................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GATGTCGATCTATGGTTTCT..................................................................................................................................................... | 20 | 3 | 7 | 0.86 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TCAGCGCCCTACTTGACGGGT.......... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TGCGGTCTCTCGAGGTCAAAAT.............................................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AGGCGTCAGGGTCGACCGT................. | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................GTCTATCGAGGTCGATAAACA........................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GGGGTCTCTCGAGGTCGAAAT.............................................................................................................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GGGTCTCTCGAGGTCAATAT.............................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AGGAGTCAGGGTCGACCTT................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................GATGTCGATTTATGGTTTCT..................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................AGTATGTTGGGGTCTTTCG........................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................AGGAAAACTCTATTGAAGA............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:6478852-6479102 + | dvi_18677 | CTTACTAATTTTTGGCTTAAATCTCATATTATTAGATAATATTTTATTTATATACATTATATCATCAGGCAACGCCAGAAAGCTCCAGCTATATATCAAAGAAGTTATACATATTTCATTTAGTTCTTTTTATATAACTTCTTGTTAATATATATTTATTGGGATATGCGTGATAAACCAACAAATTTTGTTTCATTTCAGATTTCCACGTATTTTCCGCAGTCGCAGGTGGAACTGCCCAAAACCTCGTC |
| droBip1 | scf7180000395543:505992-506030 - | CCCACTA-TTTTTAGCAAAAAGATTAGATTAAAAACTAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/16/2015 at 08:38 PM