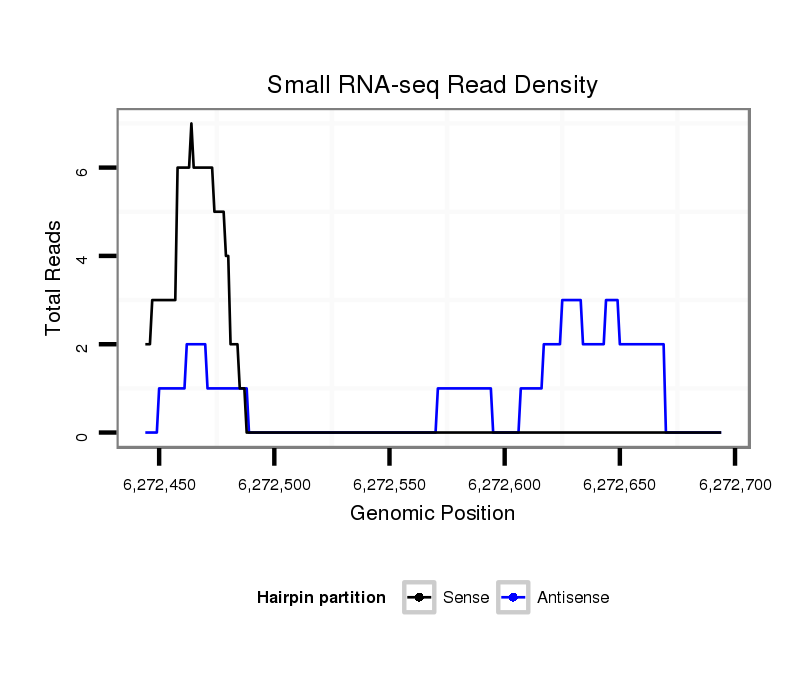
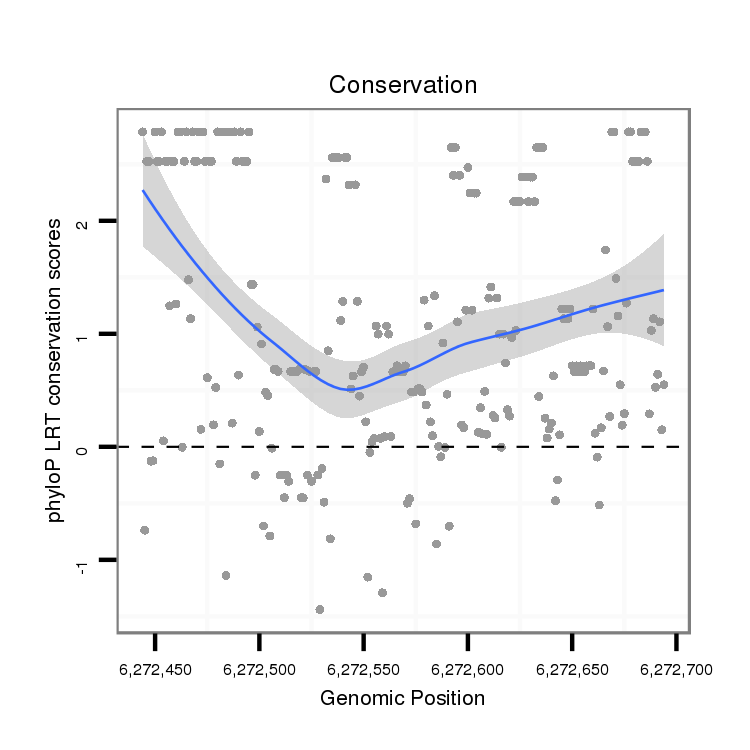
ID:dvi_18624 |
Coordinate:scaffold_13049:6272494-6272644 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_12982:1]; CDS [Dvir\GJ13061-cds]; intron [Dvir\GJ13061-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TACGTTTGGTCGGTGGCAACGATTTGGATACGCCCCTAATCATCACCAGGGTAAAGTGGAGCGAACTTCTTGCCCATCAAGTGGGGCTGCATTATGTAACCAGAGCAGATGCAGCCAAGAGCTGGCAACGGGTTCCTTCCTTCTCGCCTGTCGCTTTGTGGTTAATGTCGCCGCAACGGCCCATTCATCAGAATAAGCGACAGAGAACTGTCCATTATCGTTGCCAAACTGCTTTGGCCTTTGGGTTCTGC **************************************************...(((.((((.((((((.(((((.....(((..(((((((((.(((.......))))))))))))....)))))))).))))))))))))).......(((((((..((((.((((..((((...)))).)))).))))....)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
M047 female body |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060684 140x9_0-2h_embryos_total |
M027 male body |
SRR060667 160_females_carcasses_total |
SRR060665 9_females_carcasses_total |
M028 head |
SRR060679 140x9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060680 9xArg_testes_total |
SRR060659 Argentina_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060660 Argentina_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
SRR060678 9x140_testes_total |
SRR1106714 embryo_2-4h |
M061 embryo |
SRR060663 160_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR1106716 embryo_4-6h |
GSM1528803 follicle cells |
V053 head |
SRR1106713 embryo_0-2h |
SRR1106717 embryo_6-8h |
SRR1106719 embryo_8-10h |
SRR1106720 embryo_8-10h |
SRR1106722 embryo_10-12h |
SRR1106724 embryo_12-14h |
SRR1106726 embryo_14-16h |
SRR1106728 larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................CAACGGGGCCTTTCCTTCT........................................................................................................... | 19 | 3 | 8 | 183.50 | 1468 | 395 | 333 | 249 | 128 | 97 | 53 | 39 | 54 | 11 | 26 | 19 | 0 | 8 | 13 | 9 | 11 | 1 | 4 | 7 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGTCCTTTCCTTCTGGC........................................................................................................ | 21 | 3 | 1 | 3.00 | 3 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGTCTTTCCTTCTGGC........................................................................................................ | 21 | 3 | 1 | 3.00 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCCTTCCTTCTGGC........................................................................................................ | 22 | 3 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCCTTCCTTCTG.......................................................................................................... | 20 | 3 | 2 | 3.00 | 6 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCAACGATTTGGATACGCCCCT...................................................................................................................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCTTTCCTTCTCGC........................................................................................................ | 22 | 3 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCTTTCCTTCTCG......................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGTCTTTCCTTCTGG......................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TACGTTTGGTCGGTGGCAACG...................................................................................................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCTTTCCTTC............................................................................................................ | 18 | 3 | 20 | 1.75 | 35 | 5 | 3 | 1 | 1 | 7 | 1 | 0 | 1 | 9 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................CAACGGGGCCTTTCCTTCTC.......................................................................................................... | 20 | 3 | 2 | 1.50 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GATTTGGATACGCCCCTAATC.................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GTTTGGTCGGTGGCAACGATTTGGATA............................................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCCTTCCTTCTGG......................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGCCCTTCCTTCTGG......................................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGTCTTTCCTTCT........................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGCCCTTCCTTCT........................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGTCTTTCCTTCTGGC........................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGTCCTTTCCTTCTGGCC....................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................ATTTGGATACGCCCCTAATCATC............................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCAACGATTTGGATACGCCC........................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGTCTTTCCTTCTGG......................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGTCCTTTCCTTCTGGCC....................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGTCCCCTCCTTCTGGCC....................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTCATCTGAATAAGC..................................................... | 15 | 1 | 18 | 0.67 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| ..............................................................................................................................AACGGGGCCCTTCCTTCTGGC........................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGGTCGGTGGTAACGATTA.................................................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................ACTCCTTTGGCCGTTGGGT..... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGCCCTTCCTTCT........................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGTCTTTCCTTCTG.......................................................................................................... | 20 | 3 | 15 | 0.40 | 6 | 1 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGACCTTTCCTTCT........................................................................................................... | 19 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACGGGGCCCTTCCTTCTGGC........................................................................................................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GCAACGGGGCCTTTCCTTCT........................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CTGTCGATTAGTGGTTAA...................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................ACCCAGAGCAGATGAGGCCAA..................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................CGGGGCCCTTCCTTCTGGC........................................................................................................ | 19 | 3 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGCCCTTCCTTCTG.......................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACGGGGCCCTTCCTTCTGG......................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CCAACGGGGCCCTTCCTTC............................................................................................................ | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGTCCTTTCCTTCTG.......................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................GATGAAGGTCGCCGCAACGG........................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGCCTTTCCTTCTC.......................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AACGGGGTCTTTCCTTCTG.......................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AAAGTGGAGCGAACTGGT..................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................AATTAACACCAGGGGAAAGT.................................................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGGGACTTTCCTTCT........................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TGGGGCAGCATTATGTATAC...................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAACGGTTCCTTTCCTTCT........................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ATGCAAACCAGCCACCGTTGCTAAACCTATGCGGGGATTAGTAGTGGTCCCATTTCACCTCGCTTGAAGAACGGGTAGTTCACCCCGACGTAATACATTGGTCTCGTCTACGTCGGTTCTCGACCGTTGCCCAAGGAAGGAAGAGCGGACAGCGAAACACCAATTACAGCGGCGTTGCCGGGTAAGTAGTCTTATTCGCTGTCTCTTGACAGGTAATAGCAACGGTTTGACGAAACCGGAAACCCAAGACG
**************************************************...(((.((((.((((((.(((((.....(((..(((((((((.(((.......))))))))))))....)))))))).))))))))))))).......(((((((..((((.((((..((((...)))).)))).))))....)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060675 140x9_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
M047 female body |
SRR060658 140_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........CCACCGTTGCTAGAACTA.............................................................................................................................................................................................................................. | 18 | 2 | 6 | 3.00 | 18 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................GTAAGTAGTCTTATTCGCTGTCTCT............................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......ACCAGCCACCGTTGCTAAACC................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TCTCTTGACAGGTAATAGCAACGGT......................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GTCTCTTGACAGGTAATAGCAACGGT......................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCCCAAGGAAGGAAGAGCGGACA.................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TGCTAAACCTATGCGGGGATTAGTAGT.............................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................GTTGCCGGGTAAGTAGTCTTATTCGCTG.................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TTACAGCGGCGTTGCCGGGTAAGTAGT............................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TAGCCACCGTTGCTAGAACTA.............................................................................................................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................CAAGGAAGGAAGAGGGTA...................................................................................................... | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGAAGGAAGAGTGGAAAG................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........TAGCCACCGTTGCTAGAACT............................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................AGCAAAGGTTTGAGGAAAGC.............. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................CGTCTAAGTCGCTACTCGAC............................................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:6272444-6272694 + | dvi_18624 | TACGTTTGGTCGGTGGCAACGATTTGGATACGCCCCTAATCATCACCAGGGTAAAGTGGAG-----------CG-------AACTTCTTGCCCATCAAGTGGGGCTGCATTATGTAACCAGAG--------------------------------------CAGAT-----GCAGC---------CAAGAGCTGGCAACGGGTTCCT-----TCCTT------------------------------CTCG-----------------------------------CCTGTCGCTTT------------GTGGTTAAT-GTCGCCGC---------------AACGGCC------CATTCATCAGAAT-AA----------------------------------------------GCGACAGAGAACTGTCCATTATC---------GTTGCCAAACTG-CTTTGGCCTTTG-GGTTC--------TGC---------------------- |
| droMoj3 | scaffold_6680:4585647-4585940 + | TACGTTTGGTCGGTGGCAACGATTTGGATACGCCGTTAATCATCACCAGGGTAATGTGAAGAGTGTGGGATTTG-------------------------------TGCGTTATGTAACTAGAG--------------------------------------CAGCA-----GCAGCTGAAGAACTGAAGAGCTGGCAC--------TGTTGCTCCTCTCGCTGCTCTCCGCT------------------------------CGCTGCTCGTTGCTCTGCTGGTCGCTTGTCGCTTT------------GTGGTTGAATGTCGCCTC---------------AACGGCC------CATTCATCACAATTGTTGCACAGA------------G--------C-------G------GCGCGGCAGAGAACTGTCCATTATC---------GTTGCCAAACTG-TTTTGGCCTTTG-GGTTC--------TGC---------------------- | |
| droGri2 | scaffold_15072:83156-83445 + | dgr_231 | TCCGTTTGGTTGGTGGTAACGATTTGGATACGCCGTTAATCATCACTAGGGTAAAGT------------T-GTG-----------------------------ACTG-ATTATGTAACTAGATCAAAAACAGCCGCAGAAGAATCAACAGCAGCAAC--AGCAGCA-----GCAGC---------AAAGAGCTGGCAATGGTTTCCT-----TCCT-------------------------------CTCG-----------------------------------CCTGTCGCTTTGTCTTT------GTGGCTAAT-GTCGCCTCATCCC-----------CAGGCC------CATTCATCACAATTGTTGCACAGA---------------------A----------------GCGACAGAGAACTGTCCATTATTTGCTAGCCGTTTGCCAAACCG-CTTTGGCCTTTG-CGTTC--------TGC---------------------- |
| droWil2 | scf2_1100000004511:8285987-8286252 - | TACGTTTGGTTGGTGGCAATGATTTGGATACTCCGTTGATTATAACCAGGGTAAA-----------------TAGACACACAACCCACAGCCCAGAAGGAGGAAATGC-------------------------------------------AATAATTATGTAATAGTCGTATAGT---------CTAGT-----------------------TCTGTCTCTGGTCA--GTTTTTTGGGTTTTTGGT-----------TTTTG-------------------GGTTTTTGTAGTTTT------------GTGGTTAAT-GTCACCTCCTCTCTTAATGCGGCAGCAGCC------CATTCATCAAAAA-------------------------------------------------CCC----------------------G------CTTGCCAAACTCTTTTTGGCCTTTG-CCTGC----CTTTTGC---------------------- | |
| dp5 | XR_group8:8981285-8981476 - | TCCGTCTGGTTGGTGGCAATGATCTGGACACGCCGTTGATCATCACCAGGGTAAT------------------G----------------------------------CTTATGTAACCAGAG--------------------------------------AAGGT-----T---------------------------------CTGCTGTTCGTC-----------------------------------------TTCTGCCTGTTCGTCCG-----------TCTGTCGTC-------------------CGTC------------------------TGTGGCCCGCTCTCATTCATCAAAAT-GATGCAGAGC---------------------A-------G------AAGCC----------------------G------CTTGCCAAACCG-ATTTGGCCTTTG-TGCTC--------------------------------- | |
| droPer2 | scaffold_50:224116-224304 + | TTCGTCTGGTTGGTGGCAATGATCTGGACACGCCGTTGATCATCACCAGGGTAAT------------------G----------------------------------CTTATGTAACCAGAG--------------------------------------AAGGT-----T---------------------------------CTGCTGTTCGTC--------------------------------------------TGCCTGTTCGTCCG-----------TCTGTCGTC-------------------CGTC------------------------TGTGGCCCGCTCTCATTCATCAAAAT-GATGCAGAGC---------------------A-------G------AAGCC----------------------G------CTTGCCAAACCG-ATTTGGCCTTTG-TGCTC--------------------------------- | |
| droAna3 | scaffold_13337:8016039-8016231 - | TACGCCTGGTGGGCGGCAATGATCTGGACACGCCCCTAATAATCACCAGGGTAAT---------------AACA-----------------------------CTCG-CTTATGTAACCGGAG--------------------------------------A----------------------------------------------------------------------------------------AGCCA---------------------------GCCATTTGTCGCTTT------------GTGGCGAA----------------------------------A-----------AAAAA-AACGT--------AGAAACAGAATGCAAATTCAGATGCAGGGAAAC-----TAAGAGA----------AGT---------GAAGCCAAACCG-ATTTGGCCTTTG-TGCCC--------CCT---------------------- | |
| droBip1 | scf7180000396541:176707-176904 - | TACGCCTGGTGGGCGGCAACGATCTGGACACGCCACTAATCATCACCAGGGTAAT---------------AACA-----------------------------TTCG-CTTATGTAACCGGAG--------------------------------------A----------------------------------------------------------------------------------------AGCCA---------------------------GCCATTTGTCGCTTT------------GTGGCGAA----------------------------------A-----------AAAAA-AATA--AAA-TAAAGAAACAGAATGCAAATTCAGATGCAGGGAAAC-----TAAGAGA----------AGT---------GAAGCCAAACCG-ATTTGGCCTTTG-TGCCC--------CTT---------------------- | |
| droKik1 | scf7180000302474:360737-360849 - | TTCGCCTGGTGGGCGGCAACGATCTGGACACGCCGCTGATCATCACCAGGGTAAG------------------G-------------------------------GGCGTTATGTAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------TGCCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAAGCG-ATTTGGCCTTTG---CCT-CGCTTGCCGCT--------------------- | |
| droFic1 | scf7180000453833:801179-801368 + | TCCGATTGGTGGGCGGCAACGATCTGGACACGCCGCTAATTATCACCAGGGTAAGGCGT----------TAACG-------------------------------AGCCTTATGTAACCAGAG--------------------------------------AT-AT-----G---------------------------------CCGCC-----------------------------------------GCCACT----------------------------CTTTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAAT-GATGCACAT---------------------TC-------G------CAACG----------------------GCGGAG-----GAAAAGCG-ATTTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droEle1 | scf7180000491268:63398-63590 - | TCCGCCTGGTGGGCGGCAACGATCTGGATACGCCGCTAATAATTACCAGGGTGGG--------------TAACA-------------------------------CGCCTTATGTAACCAGAG--------------------------------------AT-GT-----G---------------------------------CC--------------------------------------------GCCACT---------------------------CTTTTGTCGCCCTGTGGTGGTGGTGGCGGTCGCT------------------------TGTGGTCCAT--TCATTCATCAAAAT-AATGCACGG-----------------------------------------C----------------------GCAGGC-----GAAAAGCG-ATTTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droRho1 | scf7180000779204:265226-265433 + | TCCGTTTGGTGGGCGGCAACGATCTGGATACGCCGCTAATAATTACAAGGGTGGG--------------TAATA-------------------------------GGCGTTATGTAACCAGAG--------------------------------------AT-GT-----G---------------------------------CC--------------------------------------------GCCACTCTTT-------------------------TGTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAAT-GATGCACAA---------------------TG----GC-G------AAATG----------------------GCAGAG-----GAAAAGCG-ATTTGGCCTTTGCTGCCC-CGCT--CCGCTTTTTGTTTTTTTC-------- | |
| droBia1 | scf7180000302428:10556467-10556635 + | TCCGCCTGGTGGGCGGCAACGACCTGGACACGCCGCTGATAATCACCAGGGTAAA-----------------CG-------------------------------GGCGTTATGCAACCGGGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTGTCGCCTT------------GTGGTCGCC-GTTGC------------------TGTGGTC------CATTCATCAAAAC-GATGCACAC---------------------TC----GCTG------G---------------------------------GCTGGCAAAGCA-ACTTGGCCTTTG-TGCCC-------CCGC---------------------- | |
| droTak1 | scf7180000414445:88017-88194 - | TCCGCCTGGTGGGCGGCAACGATCTGGACACGCCGCTAATTATCACCAGGGTAAA-----------------CG-------------------------------GGCGTTATGTAACCAGAG--------------------------------------AT------------------------------------------------------------------------------------TGCG-----------------------------------ATTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAAT-GTTGCACAT---------------------TT----GCAG---TTCTAATG----------------------GCAGAA-----GAAAAGCG-ATTTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droEug1 | scf7180000409524:41875-42043 - | TCCGTTTGGTGGGCGGCAATGATCTGGATACGCCGCTAATAATCACCAGGGTAAA-----------------TG-------------------------------GGCGTTATGTAACCAGAG--------------------------------------A-----------------------------------------------------------------------------------------------------------------------------TTGTCGTTTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAAT-GATGCACAT---------------------TC-------G------AAATG----------------------GTAGAA-----GAAAAGCG-ATTTGGCCTTTG-TGCCCCC-----TCGC---------------------- | |
| dm3 | chr3L:8619561-8619728 + | TTCGTTTGGTGGGCGGCAACGATCTGGACACGCCGCTAATTATCACCAGGGTAAA-----------------TG-------------------------------GGCGTTATATAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------G-----------------------------------ACTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAATTGAGCCAGAGA---------------------A-------G------AAATG----------------------CCTGAG-----GAAAAGCG-A-TTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droSim2 | 3l:8409002-8409169 + | TCCGTTTGGTGGGCGGCAACGATCTGGACACGCCGCTAATCATCACCAGGGTAAA-----------------TG-------------------------------GGCGTTATATAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------G-----------------------------------ACTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAATTGAGCCAGAGA---------------------A-------G------AAATC----------------------GCTGAG-----GAAAAGCG-A-TTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droSec2 | scaffold_0:898162-898329 + | TCCGTTTGGTGGGCGGCAACGATCTGGACACACCGCTAATCATCACCAGGGTAAA-----------------TG-------------------------------GGCGTTATATAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------G-----------------------------------ACTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAATTGAGCCAGAGC---------------------A-------G------AAATG----------------------GCTGAG-----GAAAAGCG-A-TTGGCCTTTG-TGCCC--------CGC---------------------- | |
| droYak3 | 3L:3292870-3293030 - | TCCGTTTGGTGGGCGGCAACGATCTGGACACGCCGCTAATCATCACCAGGGTAAA-----------------TG-------------------------------GGCGTTATATAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------G-----------------------------------ACTGTCGCCTT------------GTGGTCGCT------------------------TGTGGTC------CATTCATCAAAATTGAGG---------------------------------------------------------------------------------CAATGCA-A-TTGGCCTTTG-TGCCG--------CTT----TGT-----TCTCAGCGCC | |
| droEre2 | scaffold_4784:5426690-5426870 - | TCCGTTTGGTAGGCGGCAACGATCTGGACACGCCGCTTATCATTACCAGGGTAAA--------------TAACC---A-------------------------G-AGAGTTATATAACCAGAG--------------------------------------A----------------------------------------------------------------------------------------G-----------------------------------ACTGTCGCCTT------------GTGGCCGCT------------------------TGTGGTC------CATTCATCAAAATTGAGGCACAGA---------------------C-------G------AAATG----------------------GCAGAG-----GAAAAGCG-A-TTGGCCTTTG-TGCCCCCGCCTGCCGC---------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:19 PM