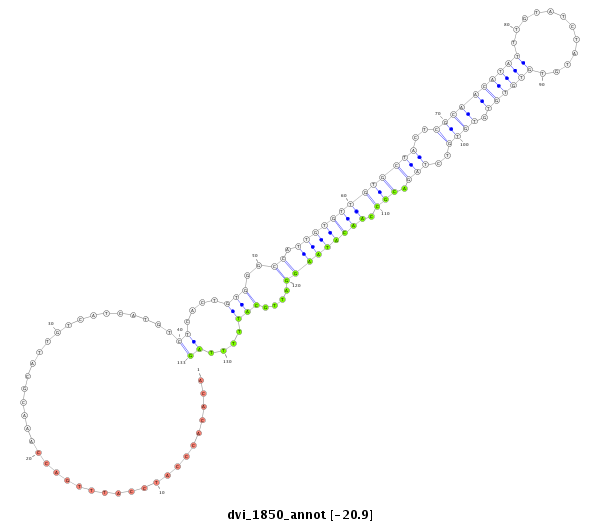
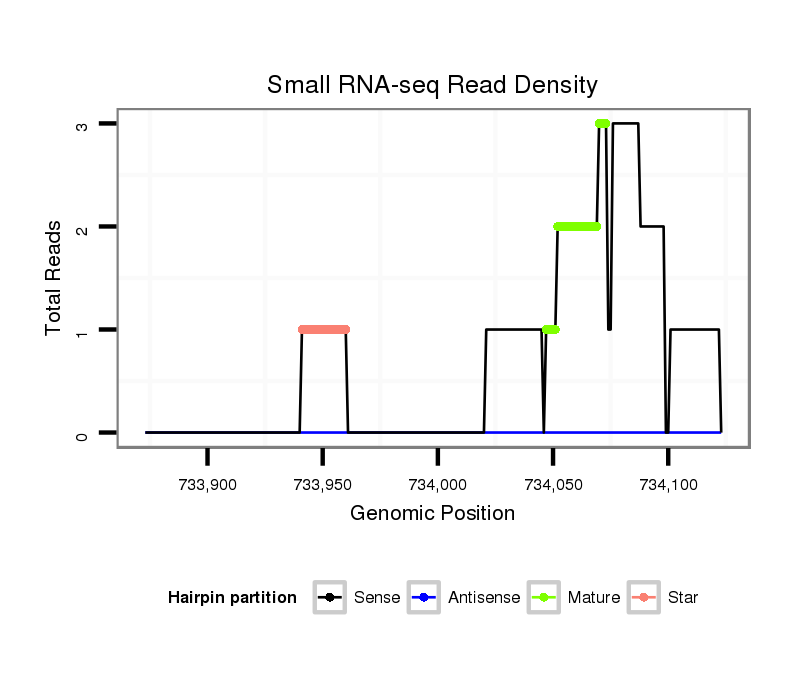
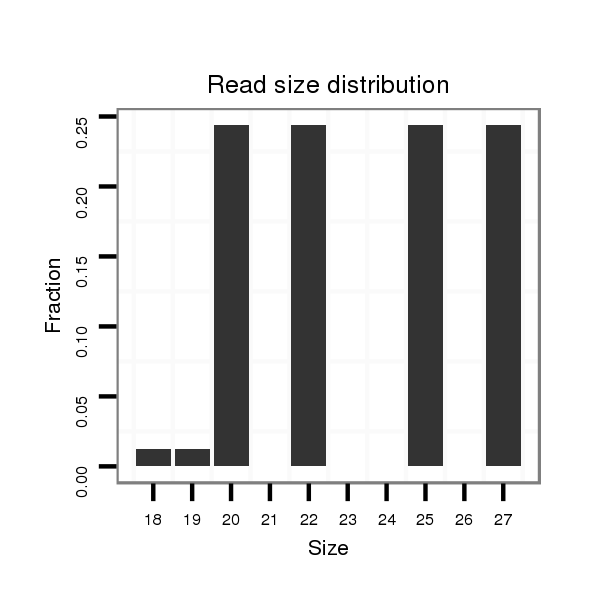
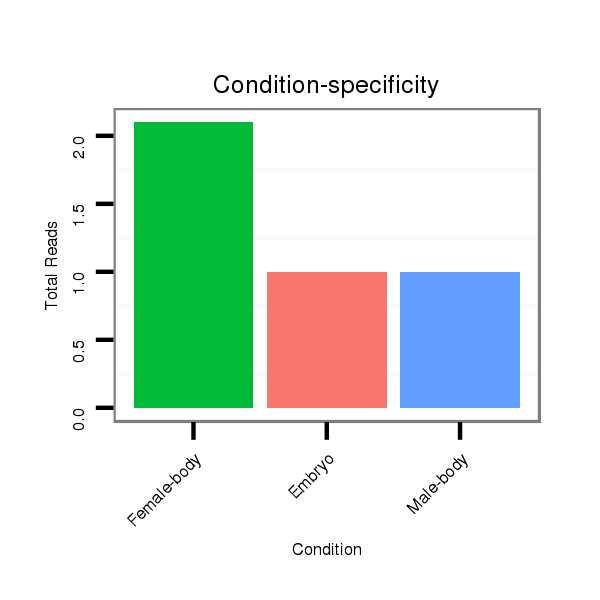
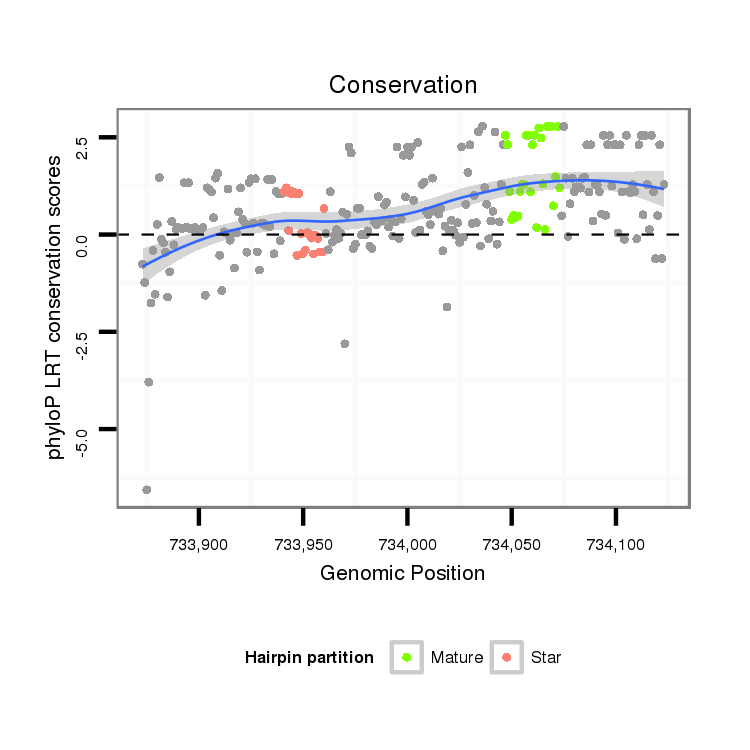
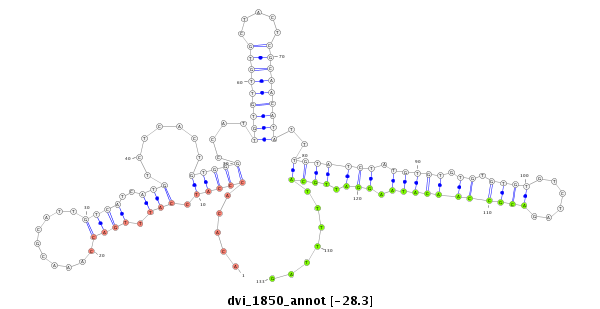
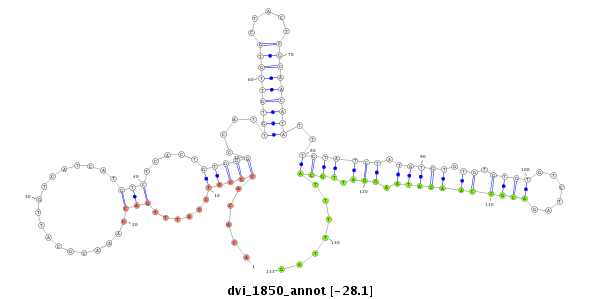
ID:dvi_1850 |
Coordinate:scaffold_12726:733923-734073 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.3 | -28.3 | -28.1 | -28.1 |
 |
 |
 |
 |
exon [dvir_GLEANR_15630:5]; CDS [Dvir\GJ15313-cds]; intron [Dvir\GJ15313-in]
| Name | Class | Family | Strand |
| (CA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATGATCACACAAATATACGCACACACACACCCACTCACCCCCACCCACTCACACACCCCCACACACTCACACACCCATCCATTTGACCAAACGCATTGTCATCATGTCTCACTGTGGGCCATTGTGTTGTGCTACTCGCAACATATTTGTATCTATGTGTGTGTGTGTGTCTAGACGCCAACATAAGGATTGCATTTTTAGATATATGGGCTTCCCCATGATCGATGCCCCAACGACAGATATATCCCGTT ********************************************************************.......................................((....(((..((.(((((((((((((..((((((((((............)))))).))))..))).))).)))))))))....)))....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060667 160_females_carcasses_total |
M047 female body |
M027 male body |
SRR060665 9_females_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
V047 embryo |
SRR060679 140x9_testes_total |
M061 embryo |
M028 head |
SRR060677 Argx9_ovaries_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................ATATGGGCTTCCCCATGATCGAT......................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TTAGATATATGGGCTTCC.................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................ACGCCAACATAAGGATTGCATTTTTAG.................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................ACACACCCATCCATTTGACC................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................AACATAAGGATTGCATTTTTAG.................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GTATCTATGTGTGTGTGTGTGTCTA.............................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CCCAACGACAGATATATCCCGT. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ATGATCACACCATTAGACG........................................................................................................................................................................................................................................ | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................CCTCATGTATCACTGTAG...................................................................................................................................... | 18 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................TATGTGTGTGTGTGTGTC................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ATCACACATATATACGGACAT................................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................CTCATGATCGAAGCCCCG................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................TCTATTTGTGTGTGTGTGTC................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................TATCTATGTGTGTGTGTGT................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TACTAGTGTGTTTATATGCGTGTGTGTGTGGGTGAGTGGGGGTGGGTGAGTGTGTGGGGGTGTGTGAGTGTGTGGGTAGGTAAACTGGTTTGCGTAACAGTAGTACAGAGTGACACCCGGTAACACAACACGATGAGCGTTGTATAAACATAGATACACACACACACACAGATCTGCGGTTGTATTCCTAACGTAAAAATCTATATACCCGAAGGGGTACTAGCTACGGGGTTGCTGTCTATATAGGGCAA
**************************************************.......................................((....(((..((.(((((((((((((..((((((((((............)))))).))))..))).))).)))))))))....)))....))******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
V047 embryo |
SRR060681 Argx9_testes_total |
SRR1106727 larvae |
M047 female body |
M027 male body |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................GTGGTACTTGCTACGGGGC................... | 19 | 3 | 4 | 1.50 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GGATTGCGTGACAGTAGTA.................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTAGTGGCTACGGGGT................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTACTTGCTACGGGGT................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTAGTTGCTACGGGGC................... | 19 | 3 | 10 | 0.70 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTACTGGCTACGGGGC................... | 19 | 2 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AGTGATACTTGCTACGGGGT................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AGTGGTACTTGCTACGGGGC................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................AGATACACACACACACACAGAT.............................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GTGGTACTGGCTACGGGGT................... | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GTGGTAGTTGCTACGGGGT................... | 19 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CGTGGTACTTGCTACGGGGT................... | 20 | 3 | 12 | 0.25 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTAGTGGCTACGGGGC................... | 19 | 3 | 18 | 0.22 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CCGGGGTAGTAGCTACGGGGT................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................ACGGGGTACTTGCTACTGGGT................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AGTTGTACTTGCTACGGGGT................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTACTTGCTACTGGGC................... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TGCCGGTCAGTGGGGGTGGGT............................................................................................................................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGTAGTAGCTACTGGGC................... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GTTGTACTTGCTACGGGGT................... | 19 | 3 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GGTTGCTGTCGCTAAAGGGC.. | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGGGGTACTGGCTACGGGGC................... | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGAGTACTGGCTACGGGGC................... | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GGTTGTTGTCTCTACAGGGC.. | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGTGGTACTGGCTACGGGGT................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CGAAGGGGTACTAGCAG......................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................GTGCTTGAGTGGTGGTGGGTG........................................................................................................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................ATGGGTGACTGGGGGGGGGTG........................................................................................................................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GTGGTACTGGCTACGGGGC................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................GTGTATTGGTGAGTGGGG.................................................................................................................................................................................................................. | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................CCCGACGGGCTTCTAGCTA......................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................GTGGTAGTAGCTACGGGGC................... | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GGGGGTAGGTGGGTGTGTGG.................................................................................................................................................................................................. | 20 | 2 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CGGGGAGAGTGTGGGGGTGTGT.......................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CGGGTACTGGCTACGGGGC................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GTGTACTAGCTACTGGGA................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GGGTGAGTGGGGCGGGGT............................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................CCGGGGTACTGGCTACGGGGT................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TCCGGGGTTGTATTCC............................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....GTGTGTTAAGATCCGTGTGT.................................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:733873-734123 + | dvi_1850 | ATGATCACACAAATATACGCACACACACACCCAC----------TCACC----------------------------------------------------------CCCA-------------CCC-----------A------C-----------TCACACACCCCC-------ACACACTCACACACCCATCCATTTGACCAAACGC-A-----------------TTGTCATCATGTCTCACT--GTGGGCC--AT--------------------------TG-TGTTGT----GCT-----------AC-T--CGCAACATAT-TTGTATCTA----------------------------------------------------------------------T--GTGT--------GTGTGTG-T-----GT---CTAGACGCCAAC-ATAAGGATTGCATTTTTAGATATATGGGCTTCCCCATGATCGATGCCCCAAC-GACAGATATATCCCGTT |
| droMoj3 | scaffold_6473:9509001-9509177 - | CACTGTT-------------------------------C---ATACATGCACGCA------------------------------------------------------------------C----------------GC-----------------ACGCACATGCACGCATC-----------------------------------T-C-----------------TCATC--C-----TTCAC--TCTGTGT-GCTC----------------------CATTGTCGTTGT---CGTT-----------GT-T---GTTG------------------------------------------------------------------------------------------------------TTGTG-T-----GT---CCAGACGTCAAC-ATAAGGATTGCTTTTTTAGATATATGGGCTTCCCGATGATCGATGCGCCAAC-GACAGATATATCCCGTT | |
| droGri2 | scaffold_14853:5168444-5168691 + | ACTATCTCACACGCACATAAATACATACATAC----------ATACATACATACA------------------------------------------------------------------T----------------A------C-----------ATACATAAATACATATCTCACAT-----CT----------------------C-A-----------------CTCACATT-----TC-AC--TGTGGGCCATT--------------------------TT-TGTTGT---GCTTGGTTTGGTTTTGTTT--TGTTGTGTGTG-TGTGTGTG--TG-------------------------------------------TCT--------G---------TCT--GTGT--------GTGTGTT-T-----GT---TTAGACGCCAAC-ATAAGGATTGCTTTTTTAGATATATGGGCTTCCCCATGATCGATGCGCCAAC-AACAGATATATCCCGTT | |
| droWil2 | scf2_1100000004510:1604775-1604958 - | ACACACACACACACACACACACACACACACACAC----------ACACA----------------------------------------------------------CACA-------------CAC-----------A------C-----------ACACACACACAC-------ACACACACACACACACACACACACACACACACAC-ATATATCTACCAAATCCCCTCTC-------------------------------------------------------------------------------AT-G--CCATGTGTGTGCCGACATTT--TG----------------------------------------------------------------------TAT--------CTTTTCC-A-----TTTCAAT------------------ATGTGTTTGTATATGTGACAACAT--------------------------------------- | |
| dp5 | XL_group1e:10474098-10474251 + | TTGGCCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAT------------------ATC--------------------------TT-TGTTGT---TGCT-----------GTTTTGTGTTGTCTGG--GGGGTGTA--TG-------------------------------------------TG--------T------------G--GTGT--------GTGTGTTGT-----GTACCACAGACGGAACC-ATAAGGACTGTGTTTTTAGGTACATGGGATTCCCGATGATCGATGCCCCCAC-GACAGACATCTCGCGCT | |
| droPer2 | scaffold_48:110612-110728 + | CACCA-------------------------------------------------------------------------------------------------------------ACACTCAT------------------------TCATACTC---A-C-------------T-----------------------------------T-C-----------------CTTTCACT-----CATCC--TTCCGCTCAATCTCTTGCCAGCTGCCAAGTGAGCCA-TC-A---------G----------------T--TGATTCGTTTTTTGTGTGTG--TG----------------------------------------------------------------------TGT--GTGT--GTGTGTC-----------------------------------------T----------------------------------------------------- | |
| droAna3 | scaffold_13047:1705064-1705279 + | dan_1160 | ACACT--------------------------------------------------------------------------CAAAAA----------------------------AAAAATCAC----------------ATATTAT-TTGGAAACAT---T-------------T-----------------------------------C-A-----------------CTTGCACC-----TTGAT--TTTGGCTCAAC--------------------------TT-TGGTGT---TGTT-----------GT-------TGTGCGTG-TGTG--TGTGGG-------------------------------------------TGTTATTTTGTGGTGTTGTGTTGTGT-------------TGTGTT-T-----GTGGTGTAGACGCAACC-ATAAGGACTGTGTTTTTAGGTACATGGGCTTCCCCATGATCGACGCCCCCAC-GACAGACATCTCCCGGT |
| droBip1 | scf7180000396740:539351-539555 + | ACACT--------------------------------------------------------------------------CA-AAA----------------------------AAAAATCAC----------------ATATTAC-CTTTAAACAT---T-------------T-----------------------------------C-A-----------------CTTGCACC-----TTGAT--TTTGGCTCAAC--------------------------TT-TGTTGTTGTTGCT-----------CT-T--TGTTGTGTGTG-TGTG--TGTGGG-------------------------------------------TGT--------T---------TTTGT-------------TGTGTT-T-----GTGGTGTAGACGCAACC-ATAAGGACTGTGTTTTTAGGTATATGGGCTTCCCCATGATCGACGCCCCCAC-GACAGACATCTCCCGGT | |
| droKik1 | scf7180000302685:801135-801260 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--TGTTGTGTGTCGTTTGTGTG--TGT---------------------------------------T--TCT--TT---------------TTGT-T----------GTGTGCG-A-----A---AATAGACGCAACC-ATAAGGACTGTGTTTTTAGGTATATGGGCTTCCCCATGATCGATGCCCCCAC-GACAGACATCTCGCGGT | |
| droFic1 | scf7180000453808:145312-145503 - | GATATTT-------------------------------CATTAT---------AATT------------T--TCACAA--------------------------------TC-GAAAATCA----------------------------------TCC-T-------------T-----------------------------------C-A-----------------TTTTCACA-----TCGATTTTTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GTTT--TGTTGTGTGA--------------------------------------------------------------------------------T--GTGT--------GTTTGTT-T-----GTGCAATAGACGCAACC-ATAAGGACTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACGGACATCTCGCGCT | |
| droEle1 | scf7180000490751:351865-352088 + | ATTACTG--------------------------------------------------------------------CACCCAAAAATCATCGATTATAAACAATTTCCTACTC-AAAAATCATCGATAACTCATTTACA-----------------TTA-A-------------T-----------------------------------C-A-----------------TTGTCATC-----ACGAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTCT--------------------------------------------------------------------------------T--GTGT--------GTTTGTT-T-----GTCCAAAAGACGCGGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTGGATGCCCCAAC-GACAGACATTTCTCGCT | |
| droRho1 | scf7180000776762:11144-11326 - | ATATT------------------------------------------------CA---------------------------CTT----------------------------AAAATTCAC-------TCATATTCA-----------------TTA-T-------------A-----------------------------------C-A-----------------TAATCACC-----TCAAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTCT--------------------------------------------------------------------------------T--GTGT--------GTTTGTT-T-----GTCCAATAGACGCGGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACGGACATCTCGCGCT | |
| droBia1 | scf7180000302187:278088-278299 - | ATTCTTGTACCATTA------------------------------------CGCACTACGCATTTCGATACTTCGCAA--------------------------------TCAAAAAACCAT---------------C-----------------TCA-T-------------C-----------------------------------T-C-----------------ATCTCATC-----TCGAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTAC--TGTGTG------------------------------------------------------------------------------T--------GTTTGTT-T-----GTGCGATAGACGCAGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCGAC-GACGGACATCTCCAGGT | |
| droTak1 | scf7180000415158:183270-183487 - | ATTGTAT------------------------------TCATTATACATTTATCCACT------------ATATGGCACTC---------------------------------AATAATCAT---------------A-----------------TTA-T-------------CC-ATA------------------------------C-A-----------------TTATCACC-----TCGAT--TTTGGCTCAAC--------------------------TT-TGTTGTTGTTGCT-----------GT-T--TGTTGTGTAC--TGTGTGTG--TG----------------------------------------------------------------------TGG--------TTTTGTT-T-----GTGCAATAGACGCAGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACGGACATCTCGCGGT | |
| droEug1 | scf7180000409458:14160-14301 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCT-----------GTTT--TGTTGTGTATTATGTGTGTG--TAT---------------------------------------G--TGT--GT--GTG--------------GTGT--------GTGTGTT-TACTATGTGGTTTAGACGCAACC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACAGACATCTCGCGCT | |
| dm3 | chrX:18802144-18802359 + | TTC------------------------------------ATTAT---------CATT------------GTATTGCAA----CAA----------------------------AAACATCAC--------CATCACCG-----------------TTA-T-------------C-----------------------------------C-A-----------------TTATCATC-----TCGAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTGTACTGCGTGTGTGTG-------------------------------------------TGT--------G---------TTT--GTTT--------GTTTGTT-T-----GTGCAATAGACGCAGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACAGACATCTCGCGCT | |
| droSim2 | x:17720070-17720272 + | ATTAT------------------------------------------------CATT------------GTATTGCAC----CAA----------------------------AAACATCAC--------CATCACCA-----------------TTA-T-------------C-----------------------------------C-A-----------------TTATCACC-----TCGAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTGTGCTGCGTGTG--TA----------------------------------------------------------------------TTT--------GTTTGTT-T-----GTGCAATAGACGCAGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACAGACATCTCGCGCT | |
| droSec2 | scaffold_8:1104964-1105166 + | ATAAT------------------------------------------------CATT------------GTATTGCAC----CAA----------------------------AAACATCAC--------CATCACCA-----------------TTA-T-------------C-----------------------------------C-A-----------------TTATCACC-----TCGAT--TTTGGCTCAAC--------------------------TT-TGTTGT---TGCT-----------GT-T--TGTTGTGTGTACTGCGTGTG--TG----------------------------------------------------------------------TTT--------GTTTGTT-T-----GTGCAATAGACGCAGCC-ATAAGGATTGTGTTTTTAGGTACATGGGCTTCCCCATGGTCGATGCCCCAAC-GACAGACATCTCGCGCT | |
| droYak3 | X:17914179-17914446 - | ATGCAAACAAACAAATACACACACCCACACACAC----------ACACA----------------------------------------------------------CACA-------------CAC-----------A------C-----------ACACACACACAA-------ACAAACACACACACTCGAATTTGCA---GAAGGCTG-----------------T------T-----AAGA----------------------------------------------------------------------T--TTTTGTGAGTTGCCTACTTG--TGTGGTCACCGAAAATATGTCTGTCTTTCGCACATAACAATGGTGTGT--G-----------------T--GTGTGTGTGTGTGTGTGTT-C-----GTTTATTTGACA---ATTTCAAAGAT---TTTTGCAGATATTT----TGCACCTGACTTGATGCACAAATAGCTGGATAAATTTGGGA | |
| droEre2 | scaffold_4690:9094379-9094607 + | GCCAC-ACATCCGTG-CCACACATCCGTGCC-ACTCTTCATTAT---------CACT------------GTATTGCAC----T-G----------------------------AAACATCGC--------C---ACCA-----------------TTA-T-------------C-----------------------------------C-A-----------------TTCTCACC-----TCGAT--TTTGGCTCAAC--------------------------TG-TGTTGT---TGCT-----------GT-T--TGTTGTGTGTACTGTG------------------------------------------------------------------------------TGT--------GTTTGTT-T-----GTGCAATAGACGCAGCC-ATAAGGATTGTGTTTTTAGATACATGGGCTTCCCCATGGTCGATGCCCCCAC-GACAGACATCTCGCGCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 11:28 PM