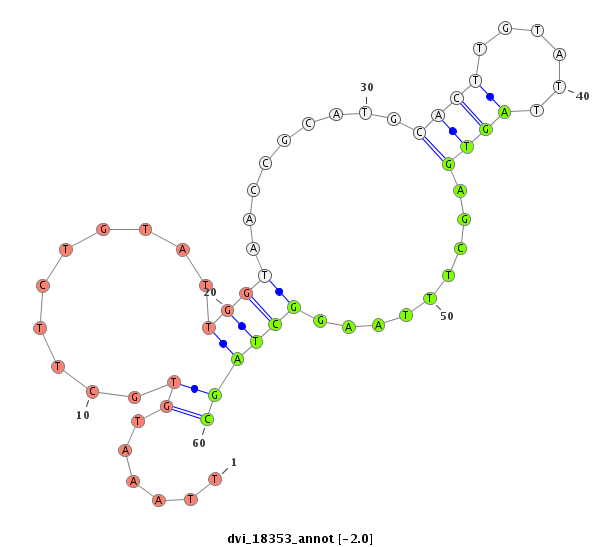
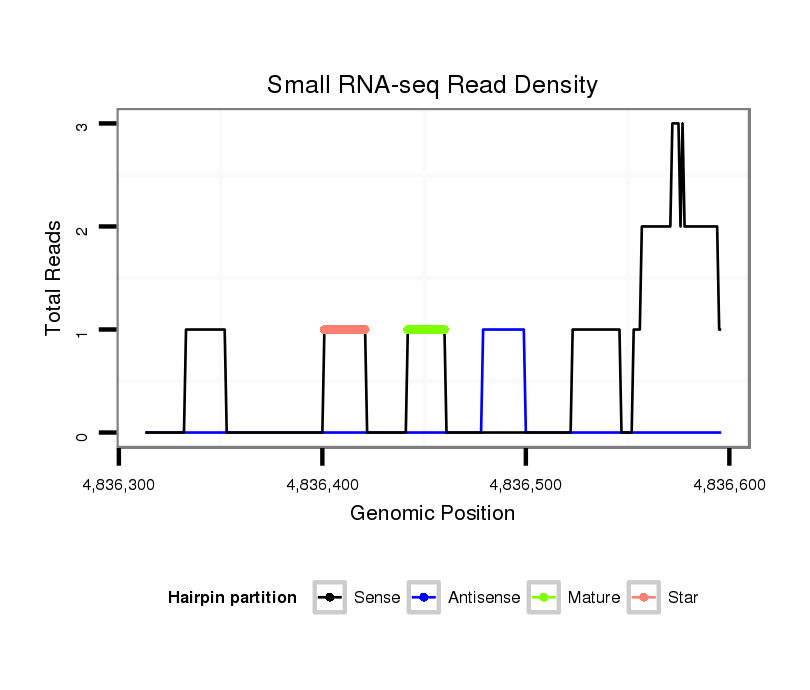
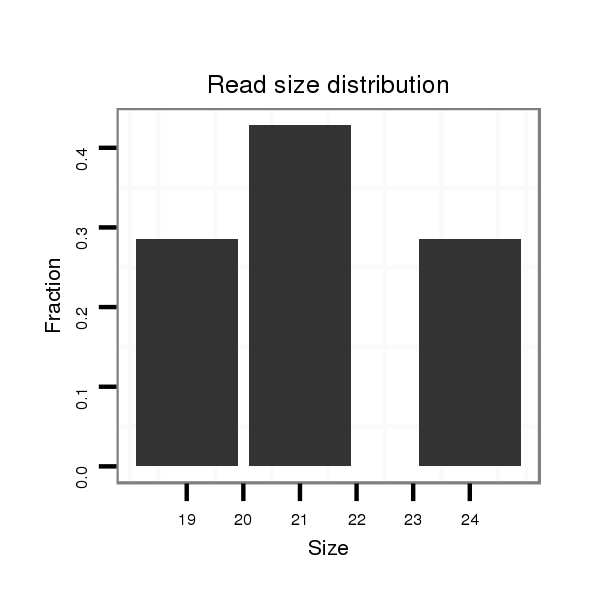
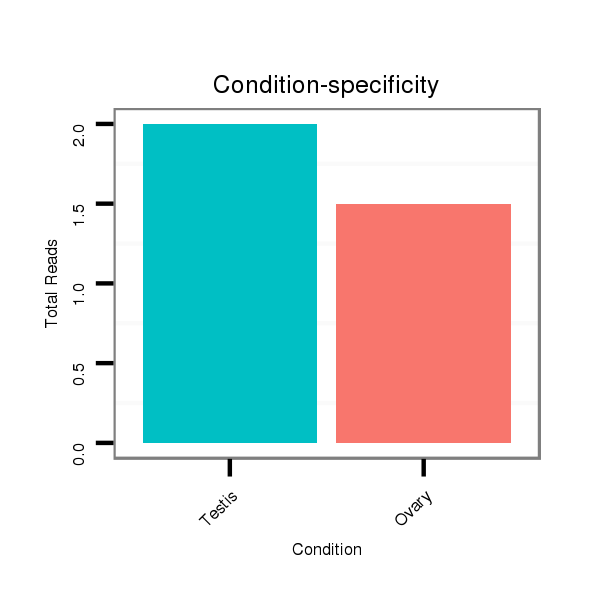
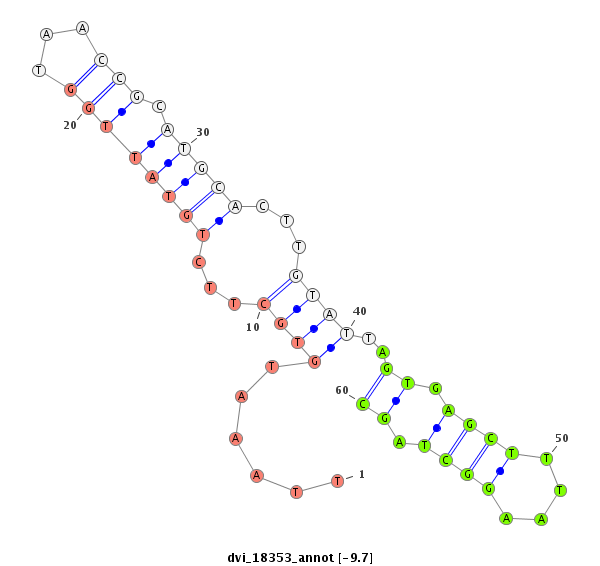
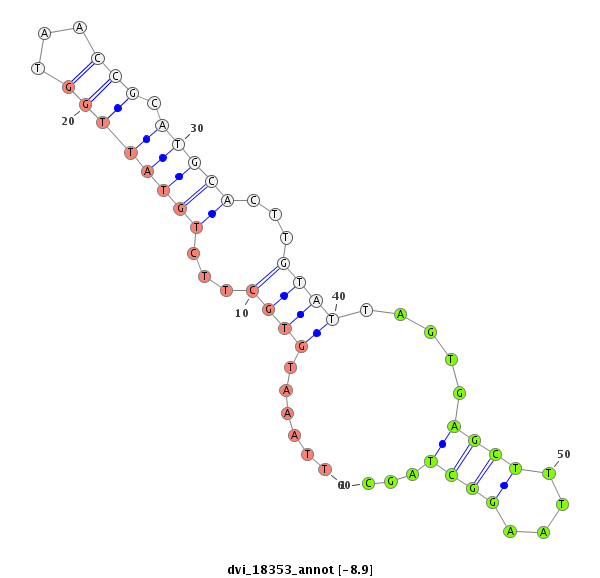
ID:dvi_18353 |
Coordinate:scaffold_13049:4836363-4836546 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
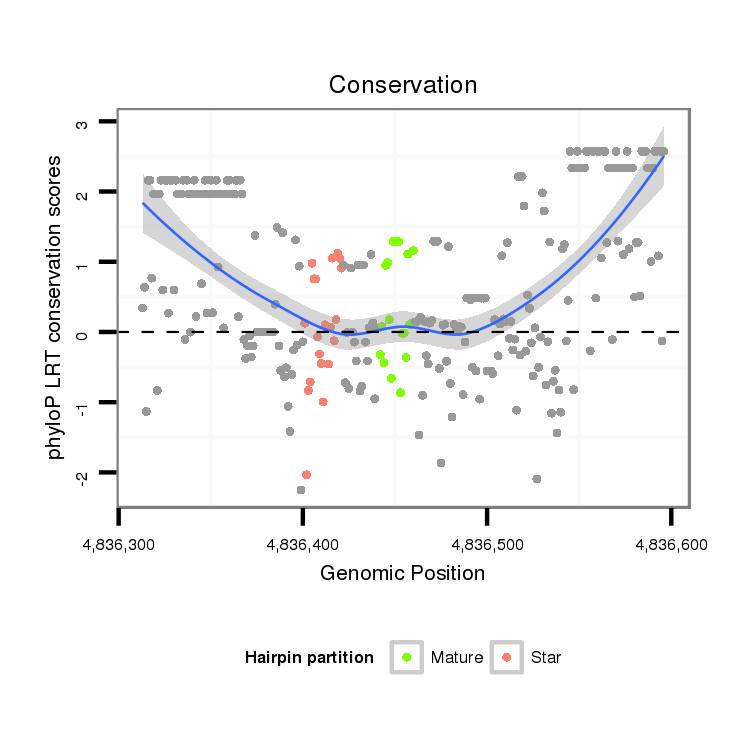
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.7 | -8.9 | -8.9 |
 |
 |
 |
CDS [Dvir\GJ12975-cds]; CDS [Dvir\GJ12975-cds]; exon [dvir_GLEANR_12904:3]; exon [dvir_GLEANR_12904:2]; intron [Dvir\GJ12975-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACAATACCGCAAAACAAATCGCTGTGCCAGGCGCTGCTCGATGGGCATCGGTAAGGACCATGATATTCGGCTAAAATGCTGCCTAAAGTTAAATGTGCTTCTGTATTGGTAACCGCATGCACTTGTATTAGTGAGCTTTAAGGCTAGCATCGCATGCATTTGCGTGTGCCTCATCCATGTTAACTTTTAAACTTATTGTAATTGTAATTGTACTTGTTTTCTCTCCGCCTTTAGCGAGGGCTTTTATTTATATCCCGATGGCCAGGCCTACAATCCGGATCTGT ****************************************************************************************......((..........((((.........((((......)))).........))))))**************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
M047 female body |
V116 male body |
SRR060687 9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
M027 male body |
SRR060681 Argx9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060659 Argentina_testes_total |
SRR060657 140_testes_total |
SRR060660 Argentina_ovaries_total |
V053 head |
SRR060663 160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060678 9x140_testes_total |
SRR060668 160x9_males_carcasses_total |
M028 head |
SRR060665 9_females_carcasses_total |
SRR060670 9_testes_total |
SRR1106729 mixed whole adult body |
SRR060655 9x160_testes_total |
SRR060664 9_males_carcasses_total |
SRR060689 160x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060676 9xArg_ovaries_total |
V047 embryo |
SRR060677 Argx9_ovaries_total |
M061 embryo |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060683 160_testes_total |
SRR1106727 larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................AGCTTTAGGGCTAGCCTCGGA.................................................................................................................................. | 21 | 3 | 3 | 90.67 | 272 | 96 | 1 | 59 | 29 | 21 | 2 | 16 | 13 | 4 | 6 | 2 | 4 | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 4 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCCTCGGA.................................................................................................................................. | 19 | 3 | 11 | 74.91 | 824 | 278 | 4 | 184 | 96 | 57 | 33 | 43 | 18 | 50 | 16 | 2 | 2 | 4 | 0 | 0 | 2 | 2 | 9 | 2 | 1 | 9 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCCTCGGA.................................................................................................................................. | 20 | 3 | 3 | 47.00 | 141 | 48 | 0 | 20 | 11 | 20 | 14 | 4 | 1 | 3 | 2 | 2 | 0 | 3 | 0 | 0 | 2 | 3 | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCGTCGCA.................................................................................................................................. | 19 | 2 | 1 | 13.00 | 13 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCGTCGAA.................................................................................................................................. | 21 | 3 | 2 | 12.50 | 25 | 5 | 0 | 4 | 4 | 0 | 6 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCGC................................................................................................................................... | 19 | 2 | 1 | 11.00 | 11 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCGTCGC................................................................................................................................... | 18 | 2 | 1 | 11.00 | 11 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TTTAGGGCTAGCCTCGGA.................................................................................................................................. | 18 | 3 | 20 | 10.25 | 205 | 123 | 0 | 0 | 22 | 0 | 17 | 14 | 9 | 0 | 4 | 0 | 0 | 1 | 0 | 1 | 5 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCGCA.................................................................................................................................. | 20 | 2 | 1 | 9.00 | 9 | 0 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCCTCGG................................................................................................................................... | 20 | 3 | 7 | 6.43 | 45 | 10 | 2 | 10 | 5 | 9 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCGTCGC................................................................................................................................... | 20 | 2 | 1 | 5.00 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCCTCG.................................................................................................................................... | 19 | 2 | 2 | 3.50 | 7 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCGTCGA................................................................................................................................... | 20 | 3 | 6 | 3.00 | 18 | 5 | 0 | 4 | 5 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATATGTGCTTGTGTATTGGTAACC.......................................................................................................................................................................... | 24 | 2 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCGAA.................................................................................................................................. | 20 | 3 | 4 | 3.00 | 12 | 4 | 0 | 0 | 1 | 0 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGTTAGCCTCG.................................................................................................................................... | 19 | 3 | 17 | 3.00 | 51 | 19 | 2 | 7 | 3 | 1 | 2 | 2 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCGTCGCA.................................................................................................................................. | 21 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCATCGGA.................................................................................................................................. | 21 | 2 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCG.................................................................................................................................... | 18 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCCTCGG................................................................................................................................... | 19 | 3 | 20 | 1.70 | 34 | 8 | 2 | 4 | 4 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AGTGAGCTTTAAGGCTAGC........................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TTAAATGTGCTTCTGTATTGG............................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TTTAGGGCTAGCGTCGCA.................................................................................................................................. | 18 | 2 | 3 | 1.00 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................CTTTTATTTATATCCCGATGGCC..................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCGCAA................................................................................................................................. | 21 | 3 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCCTCGCA.................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................TATTTATATCCCGATGGCCAG................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCGTCG.................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TAACTGCATGCACTTGTTTTAGCGAGCT................................................................................................................................................... | 28 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................TACTTGTTTTCTCTCCGCCTTTAG.................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................GGCCAGGCCTACAATCCGGATCT.. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAGCACCATGAGATTCGGCTAAA................................................................................................................................................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................GGCCTACAATCCGGATCTGT | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GCTGTGCCAGGCGCTGCTCG.................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATATGTGCTTTTGTATTGGTAACC.......................................................................................................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................TAGCTTTAGGGCTAGCGTCGCA.................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCCTCG.................................................................................................................................... | 18 | 2 | 7 | 0.86 | 6 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTCGA................................................................................................................................... | 19 | 3 | 20 | 0.85 | 17 | 2 | 0 | 7 | 1 | 0 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................ATGTGCTTGTGTATTGGTA............................................................................................................................................................................. | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GCACCATGAGATTCAGCTAAA................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TTGTAATTGTAATTGTACTTG.................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCATCGGA.................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGTTAGCATCGGA.................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTTAGGGCTAGCGTAGCA.................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................AGCGAGGGATCTTATTTGTAT............................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCTTCGGA.................................................................................................................................. | 19 | 3 | 12 | 0.17 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGACTAGCCTCG.................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGC........................................................................................................................................ | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TGCACTTGTATTAGTGAA..................................................................................................................................................... | 18 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TATTGGTAACCGGAT...................................................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................TTTATCTAGATCCTGATGGC...................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TTAGGGCTAGCGTCGCAA................................................................................................................................. | 18 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TGCATCAGTGAGCTTTAA............................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ATTGTACCTGTTATCTCT............................................................ | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCTTTAGGGCTAGCCT...................................................................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCCTCGTA.................................................................................................................................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTTTAGGGCTAGCCTC..................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGTTATGGCGTTTTGTTTAGCGACACGGTCCGCGACGAGCTACCCGTAGCCATTCCTGGTACTATAAGCCGATTTTACGACGGATTTCAATTTACACGAAGACATAACCATTGGCGTACGTGAACATAATCACTCGAAATTCCGATCGTAGCGTACGTAAACGCACACGGAGTAGGTACAATTGAAAATTTGAATAACATTAACATTAACATGAACAAAAGAGAGGCGGAAATCGCTCCCGAAAATAAATATAGGGCTACCGGTCCGGATGTTAGGCCTAGACA
****************************************************************************************************************************************......((..........((((.........((((......)))).........))))))**************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060676 9xArg_ovaries_total |
SRR060677 Argx9_ovaries_total |
M027 male body |
SRR060679 140x9_testes_total |
SRR060657 140_testes_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................GTAGCGCACGTAAACGCACAC.................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......GTGTTTTTTTTAGCGACACGGT............................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................ACGGAGTAGGTACAATTGAAA................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CACACGGAGTAGGTCTTATTG.................................................................................................... | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................CATGAACAAAGGAAAGGCGC....................................................... | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CTTTTGATTGGCGACACGGT............................................................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................CCTATTATACGACGTATTTC.................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................CCGATCGTAGCATAAGTAG............................................................................................................................ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:4836313-4836596 + | dvi_18353 | ACAATACCGCAAAACAAATCGCTGTGCCAGGCGCTGCTCGATGGGCATCGGTAAGGACCA------------------------------------------------------------------------------TGATATTCGGCTA-AAA--------------------------------------TGC------------------------------------------TG-CCT----AAAGTTAAATGTGCT---------------------------------------------TCTGTA-T---------------------------------------------TGGTAACCGCATGCACT---TGTATTAGT---GAGCTTTAAGGCTAGCATCGCATGCATTTGCGTGTGCCTCATCCATGTTAACTTTTAA-ACTTA-------TTGTAATTGTAATTG----------------------------------------------------------------------------------------------------------TACT---T-------GTTTTCTCTCCGCCTTTAGCGAGGGCTTTTATTTATATCCCGATGGCCAGGCCTACAATCCGGATCTGT |
| droMoj3 | scaffold_6680:3228009-3228277 + | ACAATACCCCAAAACAAATCGCTGTGTCAGGCGCTGCTCGACGGGCATCGGTAAGGAGAT-TAGG-AT--------------------------TA-----------------AGGCTAC----------------------------------------------------------------------------------------------------------------------CG-ATTAATG---CTGAGA---------------------------------------------TCAGAGAAAGTATT---------------------------------------------TGGTTAACGTTTAGATA---TGTGATAGG---GAGA------------------AACTTTTAAATATAATTTATATATGCTTAGTTTATT-CCATA-------------TACTAATTA--------------C----ATT------------------------C----------------------------TGCTT--------------------------CTCC---CCTACCCTATTT------CAACTTTAGCGAGGGCTTTTACTTATATCCAGATGGCCAGGCCTACAATCCCGATTTAT | |
| droGri2 | scaffold_15110:10093248-10093307 - | ACCATACCGCAAAACAAATCGCTCTGTCAGGCGCTGCTTGATGGCCATCGGTAAGAATTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004822:3114631-3114730 + | ATT---------------------------------------------------------CAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATACTCATATTCAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATTATAA----------------------------------------------------------------------------------------------------------TTCT---C--------AT----AATTTCTTTCAGTGAGGGCTTCTACTTGTATCCCGATGGTCAAGCCTATAATCCGGATCTTT | |
| dp5 | XR_group6:12892069-12892129 + | TCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTGCAGCGAGGGCTTCTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTGT | |
| droPer2 | scaffold_33:915751-915811 + | TCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTGCAGCGAGGGCTTCTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTGT | |
| droAna3 | scaffold_13337:7374878-7375217 - | ACAATCCCACAAAACAAATCGCTATGCCAAGCCCTACTCGATGGCCATCGGTAAGAGTTGAATTT-AT--------------------------TC--------------------ATAGATTCATAGACTATTCCCATG----------A-ACTACTTTTCTTAGAAATTATTATATTAAACCTTCTTAGCAT--------------------ATTGCAGTTTCTAAGAAAGAGTACG-A-------------------------------------------------------------ATCGATAAGATTAT-------------------------------------------------------------------------------GGTTTATAATTTGCATAAGATATATTTTCAA-------------------------AATTCCACTGAAATTGTAGTTTTGA------------------TAAGAATTTCAC-AGAACCCTCAGATAAGAGC----------------------------------------------------TTTCTAAACTATTTG--------T------CCCTTTTGCAGCGAGGGCTTCTACTTGTATCCAGATGGCCAAGCCATTAATCCGGATCTAT | |
| droBip1 | scf7180000395543:38438-38802 - | ACAATCCCACAAAATAAATCGCTATGCCAAGCCCTACTCGATGGTCATCGGTAAGAGTTT-ATAG-AA--------------------------TATT------------------------------------TCCATG----------A-ATTATATTTCTAAGGAATATTTATATTTATTATTATTATCGTGGACCCTGTTTGTTTGAATAATTTTTATTTCTTAGAAGAACTACC-A-------------------------------------------------------------TTCGATAAGATATA-------------------------------------------------------------------------------CGTTTAAAATTTGCATTAGATATATTTTAAAATGGCCTTTA---------------AATTCCTTGAAAATTCTA-TTTTGA------------------TAAGAATTTCCCTTGAATACCCAAATAAGAAC----------------------------------------TGCAACAAATAGTTTATCAAATATTTA--------T------ACTCTTCTTAGGGAGGGCTTCTACTTGTATCCAGATGGCCAAGCCATTAATCCGGATCTAT | |
| droKik1 | scf7180000302486:1130020-1130111 - | TCT---------------------------------------------------------TAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTGC-------TTGTGGCTGTAA------------------TAGGAAT-------------------------------------------------------------------------------------TC---------------------TTGATTGCAGCGAGGGCTTTTACTTGTATCCGGATGGCCAAGCCTACAATCCGGATCTGT | |
| droFic1 | scf7180000453833:607519-607748 + | ACCATCCCGCAAAACAAATCGCTGTGTCAGGCGCTGCTTGATGGCCATCGGTGAGAGATA-GTAG-AT--------------------------TATTAA-----------------TAG----------------------------------------------------------------------------------------------------------------------AA-------------------------------------------------------------------------------------------ATTGGA----------------AGAATTGGTGGTCTTCACATCCACTCTTTGAATTTGCTT---------AGGATATCAACACTCC-----------------------------------------------------------------------------------------------------------------------------------------CATTTCTTGTTTATTAATA--------AATAT---------------ATTATTTTCGATTTTCAGCGAGGGCTTTTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTGT | |
| droEle1 | scf7180000491268:264219-264506 - | ACGATCCCGCAGAACAAGTCGCTGTGCCAGGCGCTGCTCGATGGCCATCGGTGAGAGAAA-GAAG-AT--------------------------TGTTATTGTCTATCTAAAAAT-----------------TAACCTTG----------G-A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTA-----AGCTTAAAACACATTTGGATGTTTCTTAACC----------------------------CGTGGTTCTAATTATGTATTTTTAGAGG-------------TTTT-------------AATCGCGACTGGTTGGGAATGTTCGTTAATTCGTTTCTTGTTTACTAATA--------AAGAT---------------ATTATTTTCCATTTTCAGAGAGGGCTTCTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTGT | |
| droRho1 | scf7180000779204:77191-77493 + | ACGATCCCGCAGAACAAGTCGCTTTGCCAGGCGCTGCTCGATGGCCATCGGTAAGAGATA-ATAT-AT--------------------------TA--------------------TTAGATT-------------CACG----------ATAAC--------------------------------------TTT------------------------------------------CCGT-----------------------------------TTTTTCTCATATCTTTATTCTAATATTAATCAA----ATGGTTTATTTTTAAAAGTTAAAATTTTACACATTTCTCAGAAATAATG---------------------------------------------------------------------------------------------------------------------------------------ATTTTACCTTATATTCCCTAA------------------------GTTCGTCCA--TATTTCTTGTTTACCAACATCTTGTTTATAAT---------------ATTTTTAACCATTTTCAGTGAGGGCTTTTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTGT | |
| droBia1 | scf7180000302428:10369043-10369098 + | ACCATCCCGCAGAACAAGTCGCTGTGCCAGGCGCTGCTCGATGGCCATCGGTGAGG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000414445:294307-294410 - | TAA---------------------------------------------------------CAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---CAACTG--------TAT------------TTCAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTAATTA----------------------------------------------------------------------------------------------------------TTAT---T--------ATTATTATGAAATTTCAGTGAGGGCTTTTACTTGTATCCAGATGGCCAAGCCTACAATCCGGATCTAT | |
| droEug1 | scf7180000409524:225429-225644 - | ACAATCCCACAGAATAAGTCGCTGTGCCAGGCGCTACTCGATGGCCATCGGTGAGATATA-GTACT-TCTTACGGTTTTCTTTAAAATTTAAATTTTAAA-------------------------------------ACG----------A-AAT--------------------------------------TAG------------------------------------------CC-TCT--------------------------------------------------------------------------------------------------------------------------------------GTTAGACTACACCTAGAGTTTTAT--------------------------------------------------------------------GGCTTATTA-------------------------------------------------------------------------------------------A--------------T---------------TTTTATTTAAACTTTCAGCGAGGGCTTTTATTTGTATCCGGACGGCCAAGCTTACAATCCGGATCTGT | |
| dm3 | chr3L:8419864-8420051 + | ACAATCCCTCAGAACAAGTCGCTGTGCCAGGCGCTGCTCGATGGCCATCGGTGAGAGATA-GTATTA----------------------------------------------GTACTAG----------------------------------------------------------------------------------------------------------------------AA-CCT----AATGTCCT---TTAGTG--------TATTTTAAGAGTGTATTCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACTGATCA----------------------------------------------------------------------------------------------------------TTAT---G--------TTT------GATTTTCAGAGAGGGCTTCTACTTGTACCCAGATGGCCAAGCGTACAATCCGGATCTGT | |
| droSim2 | 3l:8225872-8226059 + | dsi_3265 | ACAATCCCTCAGAACAAGTCGCTGTGCCAGGCGCTGCTCGATGGCCATCGGTGAGGGATA-GTAGTA----------------------------------------------GTACTAG----------------------------------------------------------------------------------------------------------------------AG-CAT----AAAGTTCT---TAAGTG--------TATTATAAGAGCGTATTCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTGATCA----------------------------------------------------------------------------------------------------------TTTT---G--------TTT------GACTTTCAGAGAGGGCTTCTACTTGTACCCAGATGGCCAAGCGTACAATCCGGATCTGT |
| droSec2 | scaffold_0:715889-716076 + | ACAATCCCTCAGAACAAGTCGCTGTGCCAGGCGCTGCTCGATGGCCATCGGTGAGGGATA-GTAGTA----------------------------------------------GTACTAG----------------------------------------------------------------------------------------------------------------------AA-CCT----TAAGTTCT---TAAGTG--------TATTATAAAAGTGTATTCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTGATCA----------------------------------------------------------------------------------------------------------TTAT---A--------TTT------GACTTTCAGAGAGGGCTTCTACTTGTACCCAGATGGCCAAGCGTACAATCCGGATCTGT | |
| droYak3 | 3L:3477981-3478189 - | ACAATCCCCCAGAACAAGTCGCTGTGCCAAGCGCTGCTCGATGGCCATCGGTGAGAAATA-GTAGT-T--------------------------TACCAAAAAATAT------ATACTTG----------------------------------------------------------------------------------------------------------------------CC-CCT----AAGGCTCT---TAGTTATTATATATTATTATAAAAGTGTATTCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTGATCA----------------------------------------------------------------------------------------------------------TTAT---G--------TTT------GACTTCCAGAGAGGGCTTCTACTTGTACCCAGATGGCCAAGCGTACAATCCGGATCTGT | |
| droEre2 | scaffold_4784:5612561-5612779 - | der_736 | ACAATCCCTCAGAACAAGTCGCTGTGCCAAGCGCTGCTCGATGGCCATCGGTGAGATATT-GTAG-AT--------------------------CATTAAAGAATATC-ATATGT-----------------TGACCACG----------A-AAC--------------------------------------TGG------------------------------------------CC-CCT----AATGTCAGATGTAATTG--------TTTTATAAAAGTGTATTCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTGATAA----------------------------------------------------------------------------------------------------------TTTT---G--------TTT------GCCTTTCAGAGAGGGCTTCTACTTGTACCCAGATGGCCAAGCGTACAATCCGGATCTGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 03:57 AM