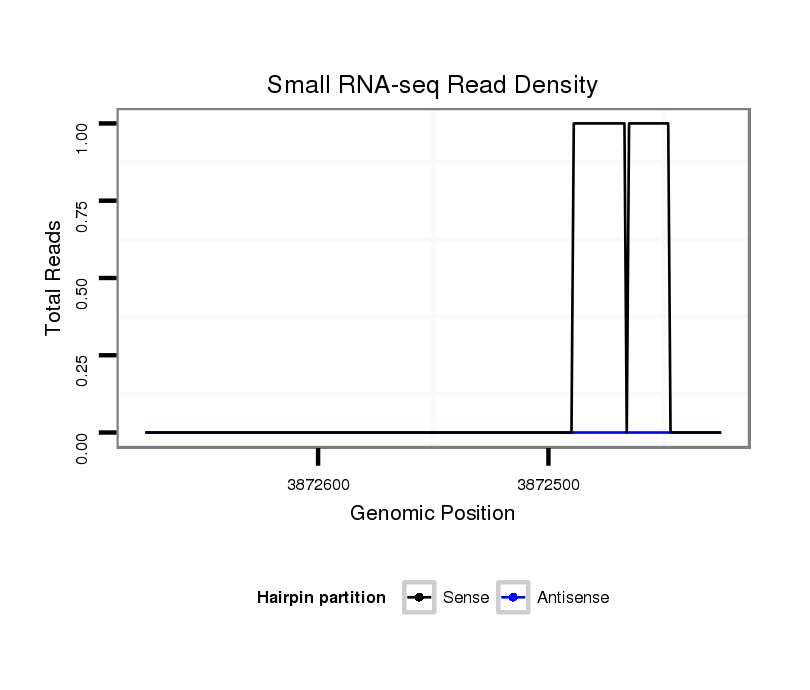
ID:dvi_18232 |
Coordinate:scaffold_13049:3872475-3872625 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_12414:2]; CDS [Dvir\GJ12446-cds]; intron [Dvir\GJ12446-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCACTTTCTGCTCAAGTTACGACTCTCTAGGATTTTACAATCATTTTTATACAATTTTAAGTACAAATAGAACAAATGGGAGTTTCAAAATTTTTTGTATATAATCTGTTAGTTTTACTATTATTCGTCGATCAGTCAGTCCCAGGCTGTGATCAGTTAAAGTTTGATTCGAGAGAAGGGAACTGCCATACTCTCTACCAGCGGCGTTCACCTGCGAGCGTGAACAGCTTTTCCGTGAGACCCTCGGACAG **************************************************.................((((....((((.((((((....(((((((.....((((......(((.(((..........(((((..(((((...))))))))))))).)))....)))))))))))..)))))).))))....))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060660 Argentina_ovaries_total |
V116 male body |
SRR060659 Argentina_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060683 160_testes_total |
SRR060689 160x9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
M027 male body |
SRR060687 9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................ATGCGAGAGTAGGGAACTGCCA............................................................... | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................ATGCGAGAGTAGGGAACTCCC................................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................CGAGAGTAGGGAACTGCCAGACA........................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CATACTCTCTACCAGCGGCGTTC.......................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CCTGCGAGCGTGAACAGC....................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TAGAATAAGTGGGAGTTTC..................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................ATACTCTCTACCAGCGGC.............................................. | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................ATGTGAGAGTAGGGAACTGCC................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TGCGAGAGTAGGGAACTGCCAG.............................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................GAGAGTAGGGAACCGCCA............................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................ACAATGATTTTTATACAAGT................................................................................................................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ACCTGAGAGGGTGAACGGC....................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGAATAAGTGGGAGTTTC..................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TCCCGTGGGTCCCTCGGA... | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ..................................................................GTAGAATAAATGGGAGCTTC..................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................CATGCGAGAGTAGGGAAC.................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CGTGAAAGACGAGTTCAATGCTGAGAGATCCTAAAATGTTAGTAAAAATATGTTAAAATTCATGTTTATCTTGTTTACCCTCAAAGTTTTAAAAAACATATATTAGACAATCAAAATGATAATAAGCAGCTAGTCAGTCAGGGTCCGACACTAGTCAATTTCAAACTAAGCTCTCTTCCCTTGACGGTATGAGAGATGGTCGCCGCAAGTGGACGCTCGCACTTGTCGAAAAGGCACTCTGGGAGCCTGTC
**************************************************.................((((....((((.((((((....(((((((.....((((......(((.(((..........(((((..(((((...))))))))))))).)))....)))))))))))..)))))).))))....))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M028 head |
M027 male body |
V116 male body |
M047 female body |
V053 head |
SRR1106729 mixed whole adult body |
SRR060671 9x160_males_carcasses_total |
SRR060667 160_females_carcasses_total |
V047 embryo |
SRR060682 9x140_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060665 9_females_carcasses_total |
SRR060666 160_males_carcasses_total |
M061 embryo |
GSM1528803 follicle cells |
SRR060683 160_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................TCGGGGTCCTACACTACTC............................................................................................... | 19 | 3 | 14 | 4.57 | 64 | 20 | 20 | 1 | 8 | 1 | 6 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 |
| ........................................................................................................................................GTCGGGGTCCTACACTACTC............................................................................................... | 20 | 3 | 3 | 3.00 | 9 | 2 | 1 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CAGGGTCCTACACTACTC............................................................................................... | 18 | 2 | 4 | 2.75 | 11 | 7 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GTCGGGGTCCTACACTACT................................................................................................ | 19 | 3 | 20 | 1.85 | 37 | 11 | 8 | 4 | 8 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGTCGGGGTCCTACACTA.................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AGATGGTCGCCTCTACTGGA...................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CTAAGAGAGCTGGTCGCCGC............................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TTCCCTTGTACGTATGAGAG........................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ACCCTCAAAATTTTA................................................................................................................................................................ | 15 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .................................................................TTAGCTTGTTTACCCACA........................................................................................................................................................................ | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CGGGGTCCTACACTAGT................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:3872425-3872675 - | dvi_18232 | GCACTTTCTGCTCAAGTTACGACTCTCTAGGATTTTACAATCATTTTTATACAATTT--------TAAGTACAAATAGAAC-AAATGGGAGTTTCAAAATTTTTTGTATATAATCTGTTAGTTT------------TACTATTATTCGTCGATCAGTCAGTCCCAGGCTGTGATCAGTTAAAGTTTGATTCGAGA------------GAAGGGAACTGCCATACTCTCTAC----------------------CAGCGGCGTTCACCTGCGAGCGTGAACAGCTTTTCCGTGAGACCCTCGGA------CAG |
| droMoj3 | scaffold_6328:4308494-4308733 - | T-------------------------CTGGGAGATTTCAATCAGTGTACTTCAAAGT--------CG-------AAGGAAA-AAATGGGTGTTTCAAAATTCGTCGTGTATAAGCTGCTACTATTAGAGTTGCATTTACAATTATTGGTCTGTCAGTCAGCCACTGGCAGTGAACACTTGGAGAATTGTAGTACGAACTGGGGCATCCAAGCCAACCACGATAAATGCCAT----------------------CAGCAAGACGCAGCTGAGATCTTTGACACCGTTAATGCGAAGACTCCGGT--------- | |
| droGri2 | scaffold_14853:705182-705432 - | T-------TCCCCAAGTTAT---ACTGTAGAAGTTTTCCGTCATACACATGTGAAATAAAAAATATTATTATTAATACAACAAAATGTGTGTTGCAAAATTTATTGTATATATTCTGTTGGTTT------------TCGAATTATACATAGAGAGGTCTATCTCAGATTCTGGCCAGACA------------------------------TGGAACTGCAGTGCCAACCATATGGAATTAATGTCCAGGAAAGAAATGACAATCGCTCATGGACTTGAAC---------TTAAGGGTTCTGGTTCTGGTTCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 09:53 PM