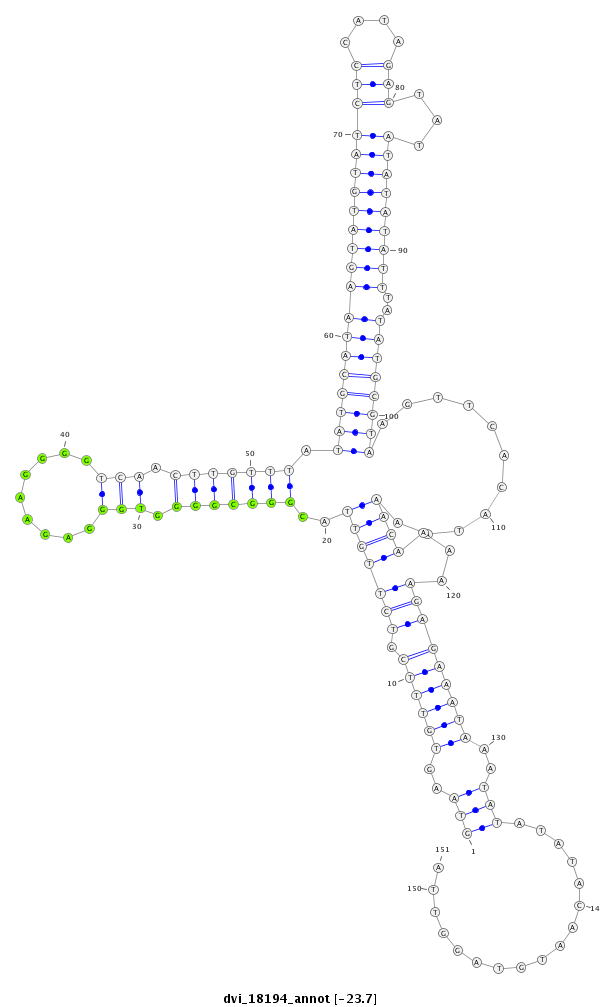
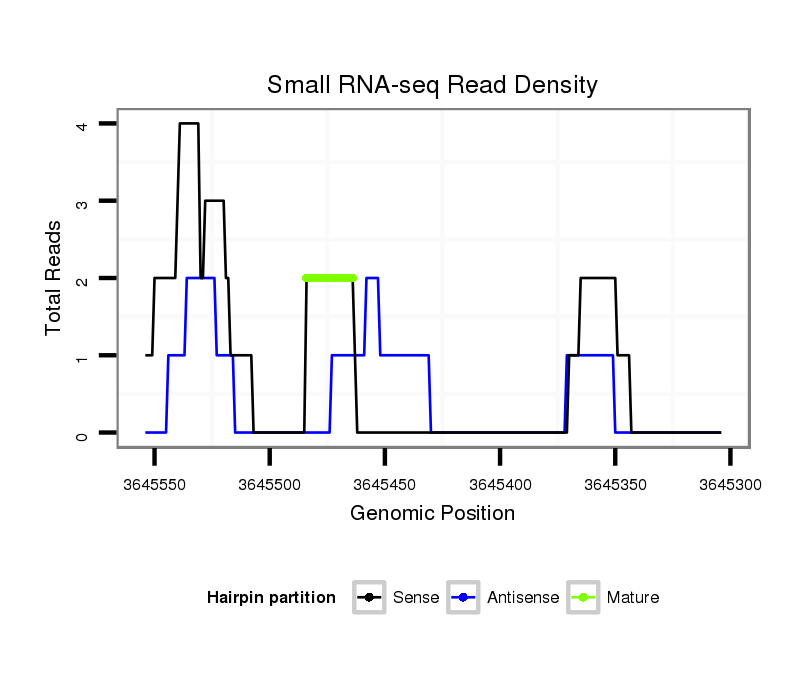
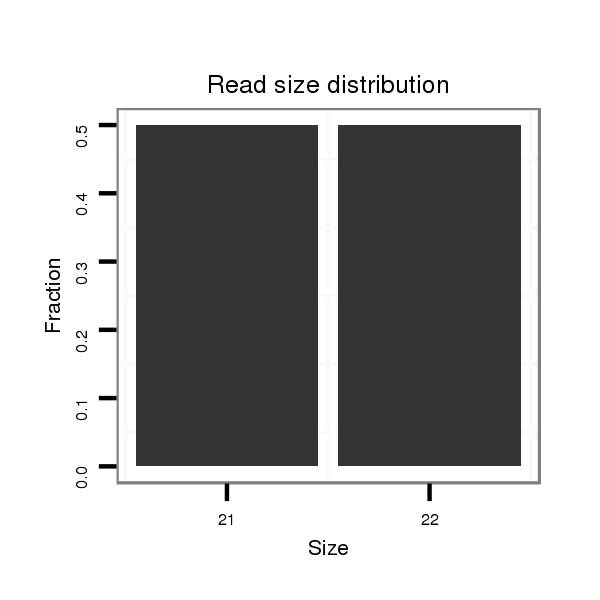
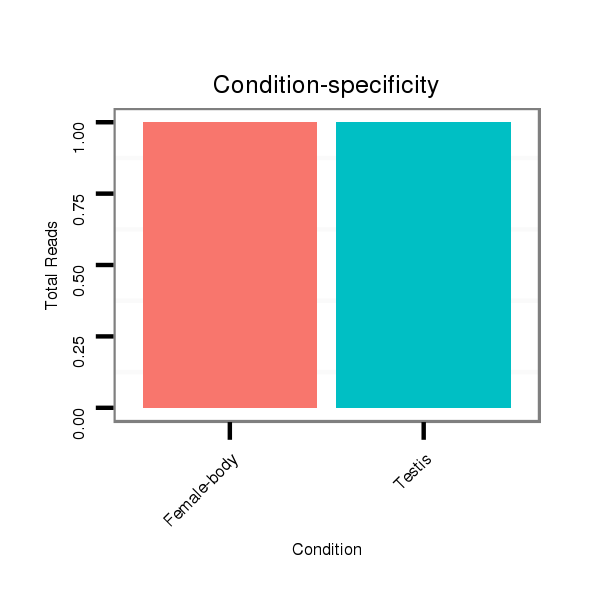
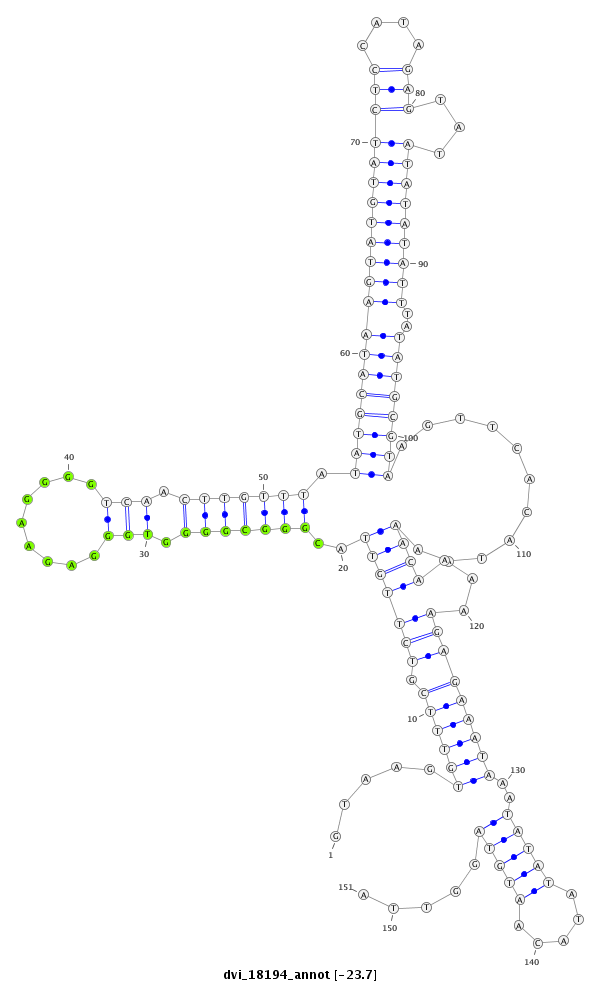
ID:dvi_18194 |
Coordinate:scaffold_13049:3645354-3645504 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.7 | -23.6 | -23.6 |
 |
 |
 |
CDS [Dvir\GJ12466-cds]; exon [dvir_GLEANR_12432:2]; intron [Dvir\GJ12466-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCGCGATATCGGCGAGATGCCAGTCACAAAGCGTTCACGCTCGGACTCGGGTAAGTGTTTCGTCTTGTTACGGGCGGGGTGGGAGAAGGGGTCAACTTGTTTATATGCATAAGTATGTATCTCCATAGAGTATATATATATTTATATGCGTAAGTTCACATAAAACAAAAAGAGAAATAAATATATATACAATGTAGGTTAATGAAACGCCAGGCCACAACAAACCTAACAAAAGGAAAATATATATATAT **************************************************(((..((((((.(((((((..(((((((.(((.........))).))))))).((((((((((((((((((((....)))...)))))))))..))))))))...........))))...)))))))))..))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060658 140_ovaries_total |
GSM1528803 follicle cells |
SRR060667 160_females_carcasses_total |
SRR060678 9x140_testes_total |
M027 male body |
M028 head |
SRR060672 9x160_females_carcasses_total |
SRR060654 160x9_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
M047 female body |
SRR060674 9x140_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
V116 male body |
SRR060663 160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR1106728 larvae |
SRR060655 9x160_testes_total |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................AAAGCGTTCACGCTCGGACTCGGAC....................................................................................................................................................................................................... | 25 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AGATGCCAGTCACAAAGCGTT........................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CACTCGGGTAAGTGTGTCGAC........................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................ACGGTCGGGGTGGGAGAAGGGGT............................................................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................CGGGCGGGGTGGGAGAAGGGG................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................CGGGCGGGGTGGGAGAAGGGGT............................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................ATATACAATGTAGGTTAATGA.............................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TAGGTTGATGAAGCGCCAGGC.................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GATATCGGCGAGATGCCAGT................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GCGCGATATCGGCGAGATGCCAGT................................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................GGACTCGGGTGGGTGTTTCGT............................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGGGTAAGTGTTTCGTCTTGTC...................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............GATGCCAGTCACAAAGCGTTCA...................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CAATGTAGGTTAATGAAACGCC........................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CAAAGCGTTCACGCTCGGACT............................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GGCGTTCACGCTCGGGCTCGG......................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TTAGAGAAGGGGTCAACTC......................................................................................................................................................... | 19 | 3 | 12 | 0.92 | 11 | 1 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................TGAATATATTTACACTGTAGGT.................................................... | 22 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGGGTGGGAGAAGGGGTT.............................................................................................................................................................. | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TTAGAGAAGGGGTCAACT.......................................................................................................................................................... | 18 | 2 | 5 | 0.40 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GCTTAGAGAAGGGGTCAACT.......................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TAGAGAAGGGGTCAACTC......................................................................................................................................................... | 18 | 3 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................GTTCACGCTCCGGCTCGGTTA...................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ATATATTTACACTGTAGGT.................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGGGTGGGAGACAGTGTCA............................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CGGGGTGGGAGACAGTGTC.............................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................GAGAAGGGGGCAACT.......................................................................................................................................................... | 15 | 1 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| GCGCGAGATCGCCGAGACG........................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AGAGATATACAATGTCGGT.................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................GGTTCGTCCTGTCACGGG................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GGGGTGGGAGACAGTGTC.............................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................CTTAGAGAAGGGGTCAACT.......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGCGCTATAGCCGCTCTACGGTCAGTGTTTCGCAAGTGCGAGCCTGAGCCCATTCACAAAGCAGAACAATGCCCGCCCCACCCTCTTCCCCAGTTGAACAAATATACGTATTCATACATAGAGGTATCTCATATATATATAAATATACGCATTCAAGTGTATTTTGTTTTTCTCTTTATTTATATATATGTTACATCCAATTACTTTGCGGTCCGGTGTTGTTTGGATTGTTTTCCTTTTATATATATATA
**************************************************(((..((((((.(((((((..(((((((.(((.........))).))))))).((((((((((((((((((((....)))...)))))))))..))))))))...........))))...)))))))))..))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060668 160x9_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
M027 male body |
SRR060671 9x160_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
V116 male body |
SRR060669 160x9_females_carcasses_total |
V053 head |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
M028 head |
SRR060689 160x9_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................ATAACTATACGAATTCAAG.............................................................................................. | 19 | 2 | 8 | 27.25 | 218 | 21 | 42 | 39 | 25 | 16 | 8 | 17 | 10 | 8 | 6 | 1 | 1 | 7 | 0 | 0 | 3 | 2 | 4 | 4 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................ATAACTATACGAATTCAAGT............................................................................................. | 20 | 2 | 3 | 9.67 | 29 | 10 | 4 | 1 | 2 | 2 | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TAACTATACGAATTCAAGT............................................................................................. | 19 | 2 | 20 | 2.10 | 42 | 9 | 0 | 0 | 4 | 6 | 6 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 1 | 3 | 0 | 1 | 0 | 0 | 2 | 0 |
| ..............................................AGCCCATTCACAAAGCAGGAC........................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................CCTCTTCCCCAGTTGAACAAA..................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CGGTCAGTGTTTCGCAAGTGC.................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CCGCTCTACGGTCAGTGTTTC............................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................ATATATGTTACATCCAATTAC............................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AACAAATATACGTATTCATACATAGAGG............................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ATAACTATACGAATTCAAGTC............................................................................................ | 21 | 3 | 20 | 0.65 | 13 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TTCCGCAGTTGAACTAGTATA................................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................CGCAAGTGCGGGGCTGAG........................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................CGGAATTGCGAGCCTGTGC.......................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TTGGTTGAATTGTTTTCCTGT............ | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................TATTGTTTTTCTCTTTAT........................................................................ | 18 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:3645304-3645554 - | dvi_18194 | GCGCGATATCGGCGAGATGCCAGTCACAAAGCGTTCACGCTCGGACTCGGGTAAGTGTTTCG--TCTTGTTACG-------------------------GGCGGGGTGG------GAGAAGGGGTCAACTTGTTTATATGCATA-----------------------------------------------------------------------------AGTATGTATCTCCATAGA---------GTATATATATATTTA----------TATGC---------------------------------------GT-------AAGTTCACATAAAA--------------------------------------------------------------------CAAA------AAG---------------AGAAATA--AATATATATACAATG--------TAGGTT--AATG------------AAAC-------------------------------------------------------------------------------------------------------------------------------GCC-----------------------AGGCCACAACAAACCTAACA--AAAGGAAA------------------------ATATAT-----A------TATAT |
| droMoj3 | scaffold_6654:403403-403644 + | GCGCGATATCGGCGAAATGCCAGCCACAAAGCGTTCGCGCTCAGACTCGGGTAAGTGTTTCG--TCTG-AT-CGT-------------------------------------------------------------------------------------GGATGCACAA--AATTCGT-------ATTAAGCATAT-----------------------AAGTATATAT--CAATTGA---------GTATATATATATAAATATATATTTATGTAC---------------------------------------GT-------AAGTTT--------------------------------------------------------------------------ATCAAA------GAGCAAGATAAATA-----TATACA--TA---------------TATATGTAGGTA--AATG------------AAAC-------------------------------------------------------------------------------------------------------------------------------GCAAGGCAA----------------CAA----------ATCAAACA--AAAGGAATATATG------------------------T-----A------TATAC | |
| droGri2 | scaffold_15110:7794097-7794350 - | GCGCGAAATCGGCGAAATGCCAGTCACAAAGAGGTCGCGTTCTGACTCGGGTAAGTGTCTTC--GCTA-TT-C-TCGCGAA----TTGCTTAT--GTACTATGTA-------------------------------------TAAGTA--TATGTG---TTTATC---TGCATAT---------------------------------------------------------------------------ATATATACATTTATATACATGTATGTAT--------GTATG-------------------------------TAGTGAGTTCATTCAAAAACAAAAGTAATACA-------------AACAAA-----ATATATGTA---------------------------------------------------------------------TGTA--------TAGGTT--AAAG------------AATC----------CACAA----------------------------------------------------------------------------------------------------------------GCAA-----------------------------------------------GGCATCAAAATG----------GAAA---ATATCT-----A------TATAT | |
| droWil2 | scf2_1100000004511:2433200-2433564 + | GCGCGAAATCAATGATATGCCGGTGACAAAAAGATCGCGCTCCGATTCAGGTGAGTCTCATATCGATG-CT--TACATTAA--------------------------------------------------------------------------------------------TTGTTT-------AAATAAAATTTACAAAAACAAAACCCTCAAAATAATATATGTAT--ACCTTCTGTGTGCATTCGATATATACTTTTATATATATAAATGTATCTTTTTTTATATATATTATTAGTCTAAAACCATAAGTATGTATGTTG---G-------AATGA-------AAAAAAAAAAATGAATC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATTATTAAAATAATATATATATATATTTACTATATATC-CTATGTCTTTGTATATAAGTTCATTTGACA----AAAAAAAAAAAAACCAATC------------------AAACGA--TAGGAAAAAAAAAAG----------AAAC--------AAAACA-AA--------- | |
| dp5 | XR_group3a:594639-594915 + | GCGCGAAATTACGGAGATGCCAGCCACAAAGCGAACGCGCTCCGACTCGGGTAAGTGTCTTG--TCCG-G-----C-------------------------------------------------------------------------------A---TGGATCTACAACAGATCCAC-------AACAGAGATAT-----------------------ATATATGTAT--GCCTTCT--------CCTATGTATATGTATACGTACACGTATGTATTATATTCAGCATG-------------------------------TACT-------------------------------------------------------------------------------------------------------------------------------------------ATGTTAGAA--ATGG------------AAACAAA-----TCAA----------ATCA----------AAGCAAAGCAAAGAAAGCGCCCCCCTCCCCCTAAGGCATATAGGCATATGCTCTCTCT--CTGTGTGTGTGTATGTACTCT------------------------------------------------------------------------------------------------------------TATAT | |
| droPer2 | scaffold_23:624699-624991 + | GCGCGAAATTACGGAGATGCCAGCCACAAAGCGAACGCGCTCCGACTCGGGTAAGTGTCTTG--TCCG-G-----C-------------------------------------------------------------------------------A---TGGATCTACAACAGATCCAC-------AACAGAGATAT-----------------------ATATATGTAT--GCCTTCT--------CCTATGTATATGTATACGTACACGTATGTATTATATTCAGCATG-------------------------------TACT-------------------------------------------------------------------------------------------------------------------------------------------ATGTTAGAA--ATGG------------AAACAAA-----TCAAATCAAATCAAATCA----------AAGCAAAGCAAAGCAAGCGCCCCCCTCCCCCTAAGGCATATAGGCATATGCTCTCTCTCTCTGTGTGTGTGTATGTACTCT---------------------------------------------------------------------------------------------TAT-----A------TATAC | |
| droAna3 | scaffold_13337:1948076-1948383 + | GCGGGAGATAAGCGAAGTCCCAACC---AAGAGATCACGCTCCGACTCGGGTGAGTTTCAAC--TCCG-TTCCGTCCCTCAGACACTAACCATACATATGGTGGGGGAGCTCTTTAACT-------------------------------AAAGGA---GGGCTC------ACCTCGTTACAAGTTA-C---------------------------------------------------------------------------------------------------------------------------------------------------AAAA----------AAAAAATAAAGAATAAGTACAAAAAACAATGAATGAATGTGTATGTATGTGTAAG----AATCTTAAGAAACTTG------------------------------GCACA--TGTATGTAGGTTTTAATC---AAATAATCAAAAAAAAACA-ATCAACAAA------ATCCACGATC-----------------------------------------------------------------------------------------------------------------------------------------------AGC-AGAGCAGCAAG----------GAGC--------CCAGCTAC------ATAA | |
| droBip1 | scf7180000396569:1991270-1991532 + | GCGGGAGATAAGCGAAGTTCCGACC---AAGAGATCACGCTCCGACTCGGGTGAGTTTCAAA--TCCG-TTCCGACCTTCAGACACAAACCAT--ATATGGCGGGGGAGCTCCTTGAAC------------------------------------A---AAGC------TCACCTCGAC-------ACC---------------------------------------------------------------------------------------------------------------------------------------------------AAAGA-------AACAAG--------------------AACAATGAATGAATGTGTACGTCTG----AG----AAT------CAACTTG------------------------------GCACATATCCATGTAGGTT--AATAATAATATATTCA-------ACA-ATCAACAAA------GTCCACGATCAGCAGAGCA-------------------------------------------------------------------------------------------------------------------------------------------AAGCAGCAAG----------GAGC--------CCGGCT------------ | |
| droKik1 | scf7180000302510:1003340-1003540 + | GCGCGAGATCAGCGACATGCCGGCCACCAAGAGATCGCGCTCCGACTCTGGTAAGTTTCCCA--TCTG-TT-CGTCTATAA----ATAACTAA--GTATGATGGA-------------------------------------ATGATA--ACCGAGGAGGCGGT----------------------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAA-----------------------------------AAC-A-----TGTTCAGTGTGT----------------------------G------------------------------GTTAA--------TAGGTC--AATC-----------A-------ACA-ATCAA--------------------------------------------------------------------------------------------------------------------------------------------------------------CAA--CATCAAGTCCCCGAGGAGCCGTAAGGAGC--------CGGCT----AA------- | |
| droFic1 | scf7180000454113:576751-576955 + | GCGAGAGATCAGCGATATGCCGGTCACGAAAAGATCGCGCTCCGACTCGGGTGAGTTTCACA--GCTG-TC-CGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATT--ACCGTG---GCGATC---AGCACATCGCC-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAAG----------------------------------AAC-A-----TGTTGTGTATGTCTG----AG----AAT------GAGCTTG------------------------------GCACA--------TAGGTT--AAGC-----------A-------GCAAATCAA--------------------------------------------------------------------------------------------------------------------------------------------------------------CA---------------AAG----------GAGC--------CGGCC----AAA-----T | |
| droEle1 | scf7180000491249:4091794-4092008 + | GCGCGAAATCAGCGACATGCCGGCCACCAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TC-CGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATA--ACCGTG---GCGATCGGCAGCACATCGCC-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAAA-----------------------------------AA-A-----TGTTGTGTATGTCTG----AG----AAT------GAGCTTG------------------------------GCACG--------TAGGTT--AATC-----------A-------ACAAATCAA----------------------------------------------------------------------------------------------------------------------------------------------------------TCATCGGA-ATCGGAATCCACGAG----------GAG--------------------------- | |
| droRho1 | scf7180000776850:454095-454306 + | GCGCGAGATCAGCGATATGCCGGCCACCAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TC-CGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATA--ACCGTG---GCGATC---AGTACATCGCT-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAA-----------------------------------AAC-A-----TTTTGTGTATGTCTG----AA----AAT------AAGCTTG------------------------------GCACA--------TAGGTT--AATC-----------A-------ACAAATCAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGAATCCACGAG----------GAGC--------CGGCA----AAA-----T | |
| droBia1 | scf7180000300910:1614962-1615026 - | GCGCGAGATCAGCGATATGCCGGCCACCAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415095:312407-312661 + | GCGCGAGATCAGCGATATGCCGGCCACCAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TC-CGTCTATAA----ATAGCTAT--GTATGATGGA-------------------------------------ATGATA--ACCGTG---GCGATC---AGCACATCGCC-------TCA---------------------------------------------------------------------------------------------------------------------------------------------------AAAA-------------AAATCA---------------AAC-A-----TGTTGTGTATGTCTG----AG----AAT------GAGCTTG------------------------------GCACA--------TAGGTT-CAATC-----------A-------AT------------------------------------------------------------------------------------------------------------------------GA---GAGCAACCAAAAACCAATCCAACAG-----------TCCACGA--AACGGAATCCACGAG----------GAGGAATCCCCACGGCCTT----------- | |
| droEug1 | scf7180000409007:320621-320847 - | GCGCGAGATCAGCGATATGCCGACCACCAAGAGATCGCGCTCCGACTCCGGTGAGTTTCCCA--GATT-TT-CGTCTATAA----ATAACTAT--GTATAGAGGA-------------------------------------ATGATA--ACCGTA---GCGATC---AGCACATCGCT-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAAA----------------------------------AAA-A----TTGTTGTGTATGTCTA----AAA---AAG------GAGCTTG------------------------------GCACA--------TAGGTT--AATC-----------A-------ACA----------------------------------------------------------------------------------------------------------------------------AAGCAA----------------CAA-----------TCTACGA--AACGGAATCCACAAG----------GAGC--------CGGCCTTCC--------- | |
| dm3 | chr3L:1501585-1501848 - | GCGCGAGATCAGCGACATGCCGGCCACCAAAAGAACGCGTTCCGATTCGGGTGAGTTTCCCA--GCTG-TT-TGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATATTGCCGCG---GCGATC---AGCACATCGCC-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAA-----------------------------------AAC-A-----TGTTGTGTGTGTCTG----GCAATGAAT------GAGCTTGACACTTACAATACATACA--TACATATGTGGCACG--------TAGGTTTCAATC-----------G-------ACA----------------------------------------------------------------------------------------------------------------------------GTGCAA----------------CAG-----------TCCACGGAATACGGAATCCTCGAG----------GAGC--------CGGCCTTCCAA------- | |
| droSim2 | 3l:1419335-1419399 - | GCGCGAGATCAGCGACATGCCGGCCACCAAAAGAACGCGCTCCGACTCGGGTGAGTTTCCCA--GCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_2:1535439-1535710 - | GCGCGAGATCAGCGACATGCCGGCCACCAAAAGAACGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TT-CGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATAGTGCCCCG---GCGATC---AGCACATCGCC-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAA-----------------------------------AAC-A-----TGTTGTGTGTGTCTG----TCAATGAAT------GAGCTTGGCACGTACAA-ACATACACCTACATATGTGGCACG--------TAGGTTTCAATC-----------G-------ACA----------------------------------------------------------------------------------------------------------------------------GTGCAA----------------CAG-----------TCCACGGAATACGGAGTCCTCGAG----------GAGC--------CGGCCTTCCAAATTGTAT | |
| droYak3 | 3L:1458382-1458626 - | GCGCGAGATCAGCGACATGCCGGCCACCAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TC-CGTCTATAA----ATAGCTAT--GTATGATGGA-------------------------------------ATGATATTGCCGTG---GCGAAC---AGCACATCGCCACAAACACCC---------------------------------------------------------------------------------------------------------------------------------------------------AAAA----------------------------------AAC-A--TATTGTTGTGTATGTCTG----AG----AAT------GAGCTTG------------------------------GCACC--------TAGGTTTCAATC-----------G-------AGA----------------------------------------------------------------------------------------------------------------------------GTGCAACCA--------------AA-----------ACCACGA--TACGGAATCCACGAG----------GAGC--------CGGCCTTCCAAA-----T | |
| droEre2 | scaffold_4784:1485634-1485885 - | GCGCGAGATCAGCGATATGCCGGCCACAAAAAGATCGCGCTCCGACTCGGGTGAGTTTCCCA--GCTG-TC-CGTCTATAA----ATAACTAT--GTATGATGGA-------------------------------------ATGATATTGCCGTG---GCGATC---AGCACATCGCC-------CCC---------------------------------------------------------------------------------------------------------------------------------------------------AAAAAAAAAAAAAAAAAA-------------AAAAAAAAACAA-----TGTTGTGTAGGTCTG----AA----AAT------GAGCTTG------------------------------GCACG--------TAGGTTTCAATC-----------G-------ACA----------------------------------------------------------------------------------------------------------------------------GTGCAACC---------------AA-----------TCCACGA--TACGGAATCCACGAG----------GAGC--------CGGCCTTCC--------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:20 PM