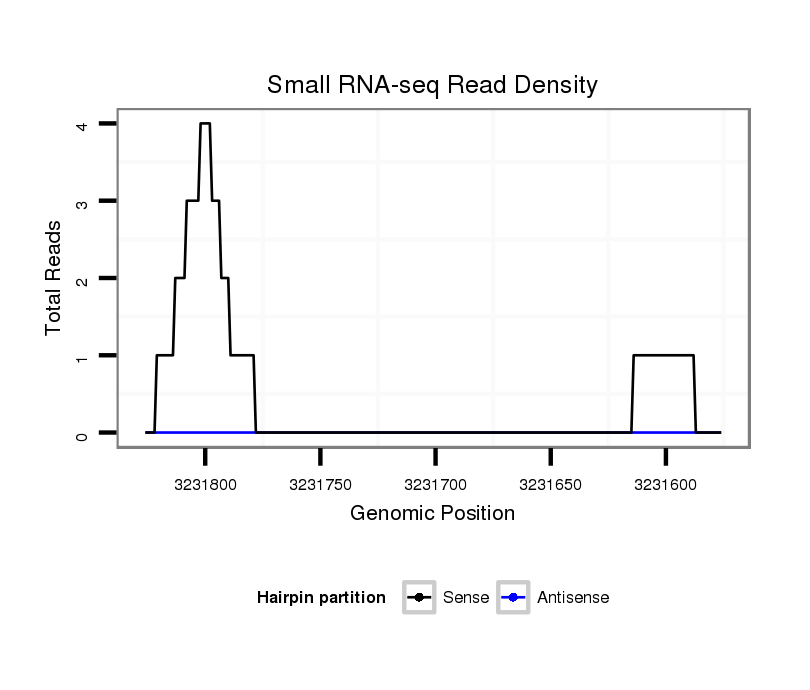
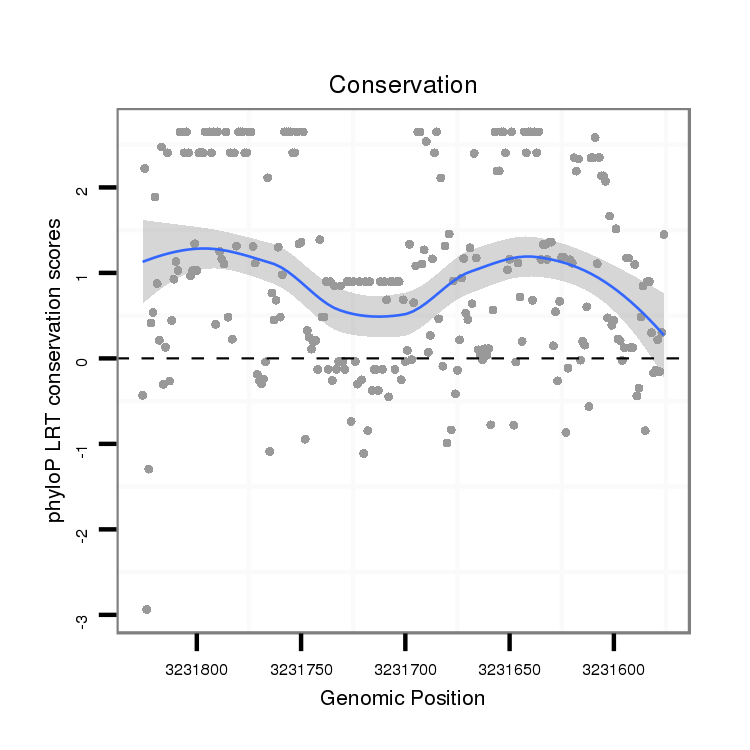
ID:dvi_18160 |
Coordinate:scaffold_13049:3231626-3231776 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ12477-cds]; exon [dvir_GLEANR_12443:5]; intron [Dvir\GJ12477-in]
| Name | Class | Family | Strand |
| (CAA)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CCAACGTTCAGGCCGAGGATCAGTCACGCGATTGTATCGCAACCCAAAAGGTAAAGTAAATGCAATACATTTGCATATATGCCAAATTTGTGCGCGCGTTCTCTTTGTATTGTTTGTTGTTGTTCTTATATGTAGCTGTTGTTGTTATTGCTATTGCTAGTTAGAATTCTGCATGCCTTCTACAGTATTGTCAACTTTTTCTTTGAGCCTAATAACCTAACCCTCGACCTGTTATTCCTTCAAAATTTGAA **************************************************(((((((((..((((((((.(((((((((.(((((((..(((((...........))))).))))).....))...))))))))).)).))))))..))))).)))).((((.((...(((...........))).....)))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060664 9_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
V053 head |
V047 embryo |
SRR060656 9x160_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................CACGCGATTGTATCGCAACCCAAA........................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CGAGGATCAGTCACGCGATT.......................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TAACCTAACCCTCGACCTGTTATTCCT............ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....GTTCAGGCCGAGGATCAGTCACGC.............................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................TAAGGCACGCGGTTGTATCG.................................................................................................................................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................ATCAGTCACGCGATTGTAT...................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CAGTCCGACGATCAGTCGCG............................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................CGGGTCATCTTTGTATTGTT......................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................GTTACTACTATTGCTAGT.......................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........CAGTCCGACGATCAGTCA................................................................................................................................................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................TCTATTTGTATTGTTAGTTGGT.................................................................................................................................. | 22 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........CAGTCCGACGATCAGCCACG............................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................TTTTTGTATTGTTTGTTGT................................................................................................................................... | 19 | 1 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................TTAGTCCCTCGAAATTTGA. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGTTGCAAGTCCGGCTCCTAGTCAGTGCGCTAACATAGCGTTGGGTTTTCCATTTCATTTACGTTATGTAAACGTATATACGGTTTAAACACGCGCGCAAGAGAAACATAACAAACAACAACAAGAATATACATCGACAACAACAATAACGATAACGATCAATCTTAAGACGTACGGAAGATGTCATAACAGTTGAAAAAGAAACTCGGATTATTGGATTGGGAGCTGGACAATAAGGAAGTTTTAAACTT
**************************************************(((((((((..((((((((.(((((((((.(((((((..(((((...........))))).))))).....))...))))))))).)).))))))..))))).)))).((((.((...(((...........))).....)))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060666 160_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060667 160_females_carcasses_total |
M047 female body |
SRR060671 9x160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060665 9_females_carcasses_total |
M028 head |
SRR060685 9xArg_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
V047 embryo |
M061 embryo |
V053 head |
SRR060657 140_testes_total |
GSM1528803 follicle cells |
SRR060675 140x9_ovaries_total |
SRR060658 140_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060678 9x140_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060676 9xArg_ovaries_total |
SRR060683 160_testes_total |
SRR060670 9_testes_total |
SRR060674 9x140_ovaries_total |
SRR060673 9_ovaries_total |
SRR1106728 larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTG...... | 20 | 3 | 5 | 127.80 | 639 | 234 | 75 | 47 | 34 | 33 | 24 | 19 | 23 | 13 | 18 | 7 | 12 | 10 | 13 | 7 | 16 | 2 | 6 | 5 | 10 | 4 | 1 | 8 | 3 | 4 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTTGAA.... | 21 | 3 | 5 | 126.20 | 631 | 335 | 64 | 17 | 28 | 23 | 26 | 23 | 11 | 14 | 21 | 7 | 12 | 6 | 1 | 6 | 5 | 4 | 4 | 4 | 1 | 3 | 4 | 1 | 1 | 4 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTTGA..... | 20 | 3 | 10 | 105.10 | 1051 | 418 | 158 | 46 | 35 | 44 | 31 | 46 | 29 | 27 | 36 | 29 | 17 | 15 | 0 | 27 | 16 | 22 | 9 | 12 | 0 | 2 | 7 | 5 | 1 | 5 | 3 | 0 | 4 | 2 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTTG...... | 19 | 3 | 20 | 96.70 | 1934 | 694 | 219 | 94 | 79 | 131 | 122 | 63 | 35 | 75 | 73 | 46 | 64 | 37 | 1 | 33 | 25 | 29 | 23 | 32 | 2 | 9 | 13 | 9 | 7 | 4 | 1 | 0 | 4 | 3 | 3 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTGA..... | 21 | 3 | 4 | 75.50 | 302 | 130 | 28 | 24 | 25 | 9 | 11 | 8 | 15 | 5 | 6 | 6 | 2 | 2 | 2 | 4 | 3 | 3 | 2 | 4 | 1 | 0 | 1 | 2 | 3 | 0 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTGAA.... | 22 | 3 | 3 | 73.00 | 219 | 121 | 14 | 10 | 16 | 9 | 3 | 10 | 7 | 4 | 0 | 3 | 1 | 1 | 5 | 4 | 2 | 0 | 2 | 1 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTT....... | 19 | 2 | 4 | 49.50 | 198 | 88 | 8 | 19 | 12 | 8 | 12 | 3 | 8 | 3 | 3 | 5 | 0 | 3 | 10 | 1 | 6 | 1 | 3 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTT....... | 18 | 2 | 8 | 44.00 | 352 | 21 | 42 | 40 | 21 | 33 | 58 | 7 | 16 | 26 | 12 | 11 | 9 | 8 | 0 | 7 | 6 | 7 | 6 | 2 | 2 | 4 | 4 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTT........ | 18 | 2 | 6 | 35.83 | 215 | 12 | 32 | 20 | 24 | 15 | 12 | 5 | 12 | 9 | 6 | 2 | 2 | 9 | 12 | 8 | 4 | 6 | 6 | 3 | 2 | 2 | 1 | 0 | 1 | 1 | 4 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GCTGGACAATAACGGAGTTTGA..... | 22 | 3 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGAAGTTTG...... | 20 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTTT...... | 19 | 2 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGTAGTTT....... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGAAGTT........ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GCTGGACAATAACGGAGTTT....... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGAAGTTTGAAG... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGAAGTTT....... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGAAGTTTGA..... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAACGGAGTTTGAAA... | 22 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTT...... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTTA..... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGTAGTTTGA..... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCTGGACAATAAGGGAGTTT....... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAAAGGAGTTTGA..... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTTTAAG... | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAAAGGAGTTT....... | 18 | 2 | 6 | 0.33 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................ACAATGAGGAAGTTTTAA.... | 18 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TATGGAAGATGTCATTACAGG.......................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACAAGTAAGTCGGATTATTG................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................ACCATGAGGAAGTTTTAA.... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAAAGGAGTTTGAA.... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TCTGGACAATAATGGAGTTT....... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GGACAATAACGGAGTTTGAAA... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAATGGAGTTTGA..... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGTTG....... | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GAAAGAAACTCGGGTTAT..................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CAGTTGAAAAAGAAATTGGGT......................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGGACAATAAAGGAGTTTGA..... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TCTGGACAATAAGGA............ | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................CAGGAAGAAACATCGACAACA............................................................................................................. | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TTGGGAGTTGTACAATCAG.............. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TTGGGAGCTTGTCAATGAG.............. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTGGACAATAACGGAGATT....... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CAGTTGAAAAAGAAATTG............................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:3231576-3231826 - | dvi_18160 | CCAA------------------------------CGT-----------------------------TC-A---------------------------------GGC-CGA-GGATCAGTCACGCGATTGTATCGCAACCCAAAAGGTAAAG--------------------------------TAAATG---------C---------------AATACATTTGCATATA-----------TGCCAAATTTGTGCG---------CGCGTTCTCTTTGTATTGTTTGTTGTTGTTCTTATATGTAGCTGTTGT----T-------------------------G---TTATTGC----TATTGC--------------TAGTTAGAATT--------------------CTGCATGC---------CTTC--TACAG-TATTGTCAACTTTT---TCTTTG---------------------------AG--CCTAA--TAACCTAACCCTCGACCT-G-TTATTCCT---TCAAAAT----TTGAA |
| droMoj3 | scaffold_6654:746256-746520 + | CCAG------------------------------CGT-----------------------------CC-AAG---------------------------------C-TGA-GGATCAGTCACGCGATTGTATCGCAACCCAAAAGGTAGGCAGAAC------------CA--------------------------------------------AATACATTTGCATATA-----------TGCCAAATTTGTGCGCAGAGTCTGCGCGTTGTCCTCCTCTAGTATGT--TTGTT--CGTTTGTAGTTGTTGT----GT----------------------------ATGTCGT----TATTGC--------------TAGTTAGAATAC--------ATAG--T--AG-TGCATG---------CCTTC--TACAG-TATTGTCAACTTAC-----TTCG---------------------------AG--CCTAAA-AAACCTAACCCTCAACCT-A-TTATTCCTAACTCCAAAA----TTGTA | |
| droWil2 | scf2_1100000004540:382992-383289 + | ACAA------------------------------GATTCA--------------CAAAAGTCGCATCAAACGTTCGA------GG---AACATCATCATCATCATCAGACGGCATCAG-GAGGCGATTGTATTAAAACTCAAAAGGTAAGG----------------------------------------------------------------AT-CATTTGCATATGCTT---AGTTATT-------TTTCCTTCGTTTT--TGTTTTCTTTTCTTTTTTGTTTTTTTTTTTTTTTTTTGTATTTTTCGT----G-------------------------G---CTATG----------AC--------TAAATTTGGTTACTGCC--------------------AT--ATGTTC------TTTCT--TATAGTTATTGTTGTTTTGT-----TTGATGTCTATTTATGCATGTGACTGCCTGCTG--CCTGCTTTTTCCT---------------------------CTAATC----TTAAA | |
| dp5 | XR_group3a:974015-974256 + | TCGT------------------------------CG--------------------CACA----------------------------TC------------TCTC-A---GGATCAGCCACGCGATTGTCTTGCATCCCAAAAGGTAAGCGATGT------------CC----CCCGGACCCCTCCTC---------CCGCCTCCTCCT---CGAG-GATTTGCATATCGTTATGCTGTATGCTGAA-------------------------------------------------TGCATGTAGTT---GTTTGTGT----------------------------TTCT----------TGTTGT---------------ACGAGTACTCGTACGAGTAG--TT--GTTGCATGCCT-CC--TCCTTC--TACAG-TATTGTCAACTTTTGTTACTTCG---------------------------AATGC--CA-TGAACCTAACCC------------------------------------- | |
| droPer2 | scaffold_23:1003887-1004138 + | TCGT------------------------------CG--------------------CACA----------------------------TC------------TCTC-A---GGATCAGCCACGCGATTGTCTTGCATCCCAAAAGGTAAGCGATGT------------CC----CCCGGACCCCTCCTC---------CCTCCTCCTCCT---CGAG-GATTTGCATATCGTTATGCTGTATGCTGAA-------------------------------------------------TGCATGTAGTT---GTTTGTGT----------------------------TTCT----------TGTTGTACGTAC-----GAGTACGAGTACTCGTACGAGTAG--TT--GTTGCATGCCT-CC--TCCTTC--TACAG-TATTGTCAACTTTTGTTACTTCG---------------------------AATGC--CA-TGAACCTAACCC------------------------------------- | |
| droAna3 | scaffold_13337:106319-106547 + | CCCA------------------------------CGTCCCCA-GCCC-------AGGCCATCCCATCGCAAAAGGAG------CC---GT------------TGCC-GAC-TGATCAGCCTCGCGATTGTCTTGCACCACTAAAGGTAAGTCAATC-------------------------GGTAATTT---------TGTTTGTTGTCT---GCGT-GATTTGCAAATCC------------------------------------------------------------------AGTC-CTAGTTGTTGT----TTG----------------ACTGTAAGT--TT----------CTTGT--------------TTG-----------------------CTTAGTTGCATGCC------GTCTTT--CATAG-TACTGTCAACTTGT--------T---------------------------AG--CCTTGCTTATCCTAACCC------------------------------------- | |
| droBip1 | scf7180000395155:650928-651140 - | CCCA------------------------------CGTCCCCA-GCCC-------GGGCCAGCCCATCGCAGAAGGAG------CC---AT------------TGCC-GAC-GGATCAGCCTCGCGATTGTCTTGCACCACAAAAGGTAAGTCAATC-------------------------GGTAGTTT---------TGTTAGCTGTCT---GCGT-GATTTGCAAATCC-------------------------------------------------------------------ATTCCTAGTTGTTGT----TTG----------------ACTTTGAGT--TA----------CTTGT--------------TAG-----------------------CTTAGTTGCATGCCTGCC--GTCTTT--CATAG-TACTGTCAACTTGT-------T--------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000297989:26436-26684 - | C--------------------------------------------------------------------AAGCGGAGTCGGAGTC---TC------------TGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCT-GGCCGCCGAATTTGAA---TC--CGAAGCCAAAT--------------------CC---AAAT-GATTTGCATATTC---CTCCGTACGT-----------------------------------------------------TGTATGTAGTTGCTGT----TTGTTTGTTTGTTTTTTTGACTTGTCGT---T-----GTTGCGATGT--------------TAG-----------------------CTTAGTTGCATGCCT------CCTCC--CACAG-TATTGTCAACTTTT-------TG----------------------------G--CCTGACCCCACCTAACCCCCAGC-------------------------------- | |
| droFic1 | scf7180000454105:565385-565639 - | GCCG-----------------------------------------------------------CATCCCAGGTGGAG------TC---TC------------CCTC-GGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCGTCC------------CA---TCC--CATGGCGGATG---------CCCC--TCGTTC---GAGT-GATTTGCATATTC------------------------------------------------------------------CGTATGTAGT-GCTGT----TTG----------------ACTTGTCGTT-CT-----GTTGAGTCGT--------------TAG------------------TCGTTCTTAGTTGCATGCCC-CC--GCCTTC--CGCAG-TACTGTCAACTTTC-------CG---------------------------AG--CCTGACCTAACCTAACCC-CAGCTCGA-CTAACCAGAGA-----AC----CCAAA | |
| droEle1 | scf7180000491249:556953-557183 - | TCGC------------------------------------AA-TCC-------CAGCAAATCGCATCCCAAGCGGAG------TC---TC------------TGCC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCATTC------------------------------------------CCCCTTTCGTCTGATGAAT-GATTTGCATATTC------------------------------------------------------------------TGTATGTAGTTACTGT----TTG----------------ACTTGTCGTTTTTGTTGT----TGTTGT--------------TAG-----------------------TTTAGTTGCATGCCC-CC--GCCTTC--CACAG-TATTGTCAACATTC-------TG---------------------------AG--CCTGACCTATCCTAACCC------------------------------------- | |
| droRho1 | scf7180000768314:9153-9385 + | G-------------------------------------------------------CAAATCCCATCCAAAGCGGAG------TC---TC------------TGTC-AGC-GGATCAGCCACGCGATTGTATTGCACCTCAAAAGGTAAGTCATCC------------CA---T-------GGTGGATC---GGAT--CCCCTTGCGTTT---GAAT-GATTTGCATATTC------------------------------------------------------------------TGTATGTAGTTGATGT----TTG----------------ACTTGTCGTT-TT-----GT-----TGT--------------TAG-----------------------TTTAGTTGCATGCCC-CC--GCCTCC--CACAG-TATTGTCAACTTTC-------CG---------------------------AG--CCTGACCTATCCTAACCCCCAGCTCG----------------------------- | |
| droBia1 | scf7180000300910:518933-519213 - | CCTGGACGAG---GAGTCCCAGTCGCATTCCCAT--TCGCCA-TCC-------CAGCACATCCCATCCCAAGCGCAG------TC---TC------------TGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCATCC------------TCCCATGGTGGATGGTAGT-----------CCCCCTGCGTCT---GAAT-GATTTGCATATGC------------------------------------------------------------------TGTACGTAGTTACTGT----TTG----------------ACTTGTCGTT-TT-----GTTGTGTTGT--------------TAG-----------------------TTTAGTTGCATGCCC-CC--GCCTCC--CACAG-TATTGTCAACTTTC-------TG---------------------------AG--CCTGACCTATCCTAACCCCCAGC-------------------------------- | |
| droTak1 | scf7180000415846:263551-263796 + | CCAG-----------------------------------------------------------CTTCCCAAGCGGAG------TCTTCTC------------TGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCATCT------------CA---TGGAGGATGCTAGATGATCCCATCCCCCGTTTCGTCT---GAAT-GATTTGCATATCC------------------------------------------------------------------TGTATGTAGTTACTGTTTGTTTG----------------ACTTGTCGTT-TT-----GTTGTGTTGT--------------TAG-----------------------TTTAGTTGCATGCCC-CCTTGCCTCCCACACAG-TATTGTCAACTTTC-------TG---------------------------AG--CC--GCCTAACCTGACC-------------------------------------- | |
| droEug1 | scf7180000409184:474485-474734 - | CCCA-----------------------TTCCCAG--TC-CCAGTCC-------CAGCCAGTCCCATCCCAAGCGGAG------TC---TC------------TCTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGTCATCC------------CA----------TGGTGGATC---------CCTCTGTCGCCT---GAAT-GATTTGCATTTTC------------------------------------------------------------------CGTATGTAGTTACTGT----TTG----------------ACTTGTCGTC-TT-----GTTGCGTTGT--------------TAG-----------------------TTTAGTTGCATGCCC-CC--GCCTTC--CACAG-TATTGTCAACTCTC-------CT---------------------------AG--CCTGACCTAGCCTAACCCCC----------------------------------- | |
| dm3 | chr3L:453927-454196 - | CCAA------------------------TCCCAG----------TCGCGGTCACAGCAAGTCCCATCCCAAGCGAAG------TC---GC------------CGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGCCATAC------------CACCACGATGGATGGTAGATG---------CCCCTTTCGTCT---GAAT-GATTTGCATATTC-------------CGTA-------------------------------------------------TGTATGTAGTTACTGTTTGTTTG----------------ACTTGTCGTT-CT-----GTTGTGTTCT--------------TAG-----------------------CTTAGTTGCATGCCC-CC--GCCTCA--CATAG-TATTGTCAACTTTC-------TG---------------------------AG--CCTGACCTATCCTAACCCCCAGC-------------------------------- | |
| droSim2 | 3l:417633-417907 - | dsi_18227 | CCTGGATGAGACC------------CAATCCCAG----------TCGCGGCCACAGCAAGTCCCATCCCAAGCGAAG------TC---GC------------CGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGCCATAC------------CA---CCTTGGATGGTAGATG---------CCCCTTTCGTCT---GAAT-GATTTGCATATTC-------------CGTA-------------------------------------------------TGTATGTAGTTACTGT----TTG----------------ACTTGTCGTT-CT-----GTTGTGTTCT--------------TAG-----------------------CTTAGTTGCATGCCC-CC--GCCTAA--CATAG-TATTGTCAACTTTC-------TG---------------------------AG--CCTGACCTATCCTAACCCCCAGC-------------------------------- |
| droSec2 | scaffold_2:475997-476243 - | CCA--------------------------------------------------CAGCAAGTCCCATCCCAAGCGAAG------TT---AC------------CGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGCCATAC------------CA---CCTTGGATGGTAGATG---------CCCCTTTCGTCT---GAAT-GATTTGCATATTT-------------CGTA-------------------------------------------------TGTATGTAGTTACTGT----TTG----------------ACTTGTCGTT-CT-----GTTGTGTTCT--------------TAG-----------------------CTTAGTTGCATGCCC-CC--GCCTCA--CATAG-TATTGTCAACTTTC-------TG---------------------------AG--CCTGACCTATCCTAACCCCCAGC-------------------------------- | |
| droYak3 | 3L:430282-430571 - | CCTGGATGAGACC---------------------CGT------TCC-------CAGCCAGTCACATCCCAAGCGAAG------TC---GC------------CGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGCCATAC------------GA---CGGTGGATGGCAGATG---------CCCCTTTCGTCT---GAAT-GATTTGCATATTC-------------CGAA-------------------------------------------------TGTATGTAGTTACTGT----TTG----------------ACTTGTCGTT-CT-----GTTGTGTTCC--------------TAG-----------------------CTAAGTTGCATGCCC-CC--GCCTCA--CATAG-TATTGTCAACTTTC-------TG---------------------------AG--CCTGACCTATCCTAACCCCCAGCTCAAGCTAACCGAAGA-----ACCTACATGAA | |
| droEre2 | scaffold_4784:451882-452159 - | CCTGGACGAGATCCA------------CTCCCAG--TC-CCA-GTCCCGGTCACAGCAAGTCCCATCCCAAGCGAAG------TC---GC------------CGTC-AGC-GGATCAGCCACGCGATTGTCTTGCACCTCAAAAGGTAAGCCATAC------------CA---CGTTGGATGGTAGATG---------CCCCTTTCGTCT---GAAT-GATTTGCATATGT-------------CGTA-------------------------------------------------TGTATGTAGTTACTGT----TTG----------------ACTTGTCGTT-CT-----GTTGTGTTCC--------------TAG-----------------------CTTAGTTGCATGCCC-CT--GCCTCC--CTTAG-TATTGTCAACTTTC-------TC---------------------------AG--CCTGACCTATCCTAACCCCC----------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:50 PM