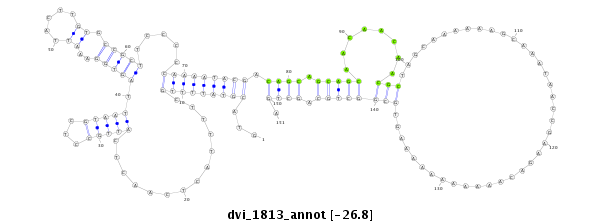
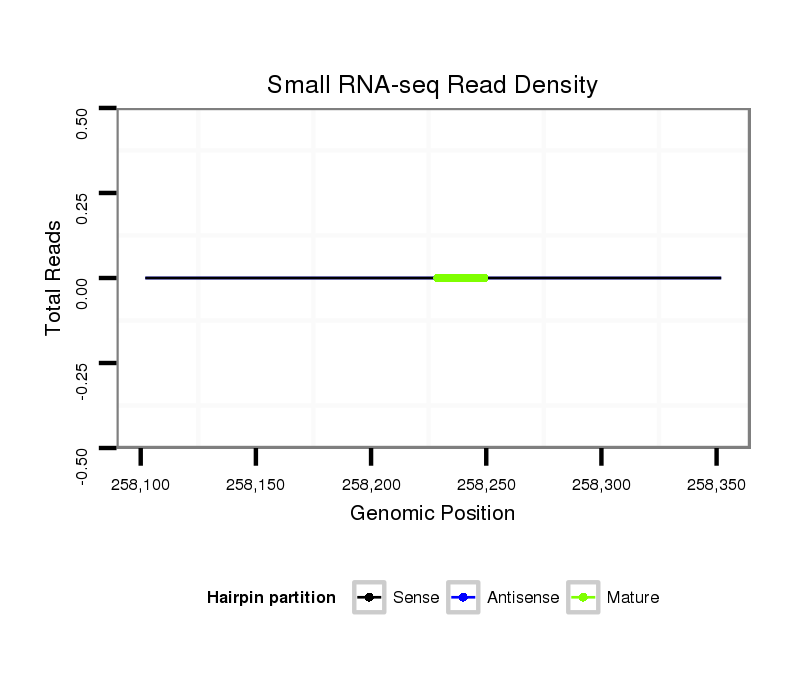
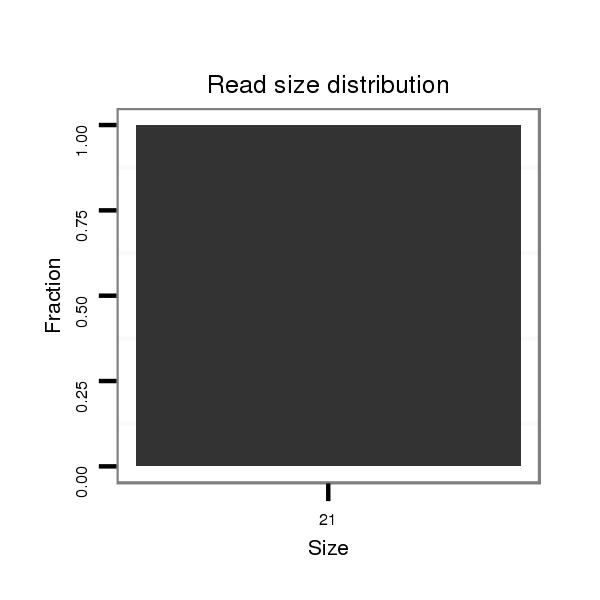
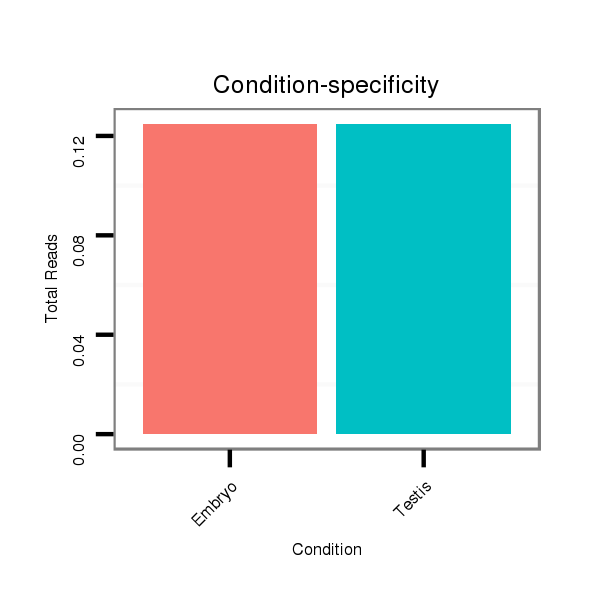
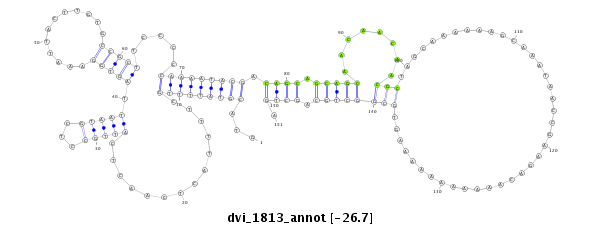
ID:dvi_1813 |
Coordinate:scaffold_12726:258152-258302 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.72 | -26.57 | -26.47 |
 |
 |
 |
CDS [Dvir\GJ15297-cds]; exon [dvir_GLEANR_15616:2]; intron [Dvir\GJ15297-in]; Antisense to intron [Dvir\GJ15274-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACCCCATGCAGATGCAAGCAACTGTTGCAGTTGCTGTTGCAGTTGCAAACGTACGTATTTTGCTTTTACTCAACTCATTGCCTCGTAATTAGTGGAAATTACTTGTGCCGCTTCCCCCAAAATACGACAGCAGCAGCAACAACAACGCTAGCAAAAAAGCAAATAACCGAAGACAAAAAAAAAAAGTGCGGCTGCAGCTGAGAGTCGCTTGTAATTACCGCAGCGATCACAGAACAACGATCAACGTCCAC **************************************************...(((((((((..............(((((...))))).(((((..((.....)).))))).....))))))))).((((.(((((........(((.......................................)))))))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060673 9_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........AAGATCCAAGCAACTGTT................................................................................................................................................................................................................................. | 18 | 2 | 18 | 0.39 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CAGCAGCAGCAACAACAACGC....................................................................................................... | 21 | 0 | 8 | 0.25 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TTACCGAAGACAACAAAAA..................................................................... | 19 | 2 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ......TGAAGATGCAAGCACTTGTTG................................................................................................................................................................................................................................ | 21 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................ACAGTCCGACGATCAACGT.... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TAACCATAGACAAAAAAAAAA................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................CAGTCCGACGATCAACGT.... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................GTCCGAAGACAAAAAAAA..................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGGGGTACGTCTACGTTCGTTGACAACGTCAACGACAACGTCAACGTTTGCATGCATAAAACGAAAATGAGTTGAGTAACGGAGCATTAATCACCTTTAATGAACACGGCGAAGGGGGTTTTATGCTGTCGTCGTCGTTGTTGTTGCGATCGTTTTTTCGTTTATTGGCTTCTGTTTTTTTTTTTCACGCCGACGTCGACTCTCAGCGAACATTAATGGCGTCGCTAGTGTCTTGTTGCTAGTTGCAGGTG
**************************************************...(((((((((..............(((((...))))).(((((..((.....)).))))).....))))))))).((((.(((((........(((.......................................)))))))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
V116 male body |
V053 head |
V047 embryo |
SRR060689 160x9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060659 Argentina_testes_total |
SRR060665 9_females_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060683 160_testes_total |
SRR060674 9x140_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
M028 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................CAACGGTAGCATGCAGAAAA.............................................................................................................................................................................................. | 20 | 3 | 13 | 1.69 | 22 | 7 | 2 | 0 | 2 | 0 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGCGTCAAAGTTGGCATGC.................................................................................................................................................................................................... | 19 | 3 | 9 | 0.33 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TTTTATGCTGCCCTCGTCGTCGT.............................................................................................................. | 23 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GTCGTGATGCTAGTTGCA.... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGCGTCAACTTTGGCATGC.................................................................................................................................................................................................... | 19 | 3 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TCACTCCGACGTCCACTATC............................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGCTTCAACGTTGGCATGC.................................................................................................................................................................................................... | 19 | 3 | 19 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ................................................................................................................................................................................................................AACATTAAGGGTGTCGATAG....................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CGATGTTGTTGCGACCGTTC................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TCTGCTGTCGTCGTCGTTGT.............................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12726:258102-258352 + | dvi_1813 | ACCCC-A-----TGCAGATGCAAGCAA---CTG---------------TTG------CAG----------------------------TTG------------------CTGTTGCAGTTGCAAACG------------------------------------------------------TA-------------------------------------------------------------------------------------------CGTATTTTGCTTTTACTCAAC------------TCATTGCCTCGTAATTAGTGGAAATTA-----------------------------------C----TTGTGCCGC-------------------------------------------------------TTCCCCCAAAATACGACAGCA------GC---A-------------------------------------------GCAA---------CAA---CAAC---GCTAGC---------------------------------------AAAAA-----AGCAAATAA-------CCGAAGACA--------A------AA-------------------------------------------------A---AAAAAAGTGCGGCTGCAGCTGAGAGTCGCTTGTAATTACCGCAGCGATCACAGAACAACGATCAACGTCC---AC----------------------- |
| droMoj3 | scaffold_6308:1910244-1910488 + | ACCCC-A-----TGCAGATGCATGCAA---CTG---------------TTG------CAG----------------------------T--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTTGCTTTTGCTCAAC------------TCATTGCCTCGTAATTAGTGGATATTA-----------------------------------C----TTGTGCCGC-------------------------------------------------------TTCCCCCAAAATCCGACAAC---AGCTTT---G---ACTGCA----------------------------------------------------------------------------------------------------------AAAA-----AGAAAAAAA-------------------------------AA----AAAAAAAAAGAAAAAACCG-GCCGAGAAGAAGCAGA--------AGA----AAAAGTGCGGCTGCAGCTGAGAGTCGCTTGTAATTACCGCAACGATCACCAAACAACGATCAACGTCCACTGC----------------------- | |
| droGri2 | scaffold_15203:2470121-2470444 + | ACCCC-A-----TGCAGATGCACGCAA---CTG---------------TTG------CAG----------------------------TTG------------------CACTTGCTGCTGCTGCTG------------------------------------------------------CTGCT---------GCT--------------------------------------------------------GCTACTGTTGCAAAGTTGTACGGATTTTACTTTTACTCAAC------------TCATTGCCTCGTAATTAGTGGAAATTA-----------------------------------C----TTGTGCCGT-------------------------------------------------------TTC-CCCAAAATGC------AACAACAAC---A---ACTGCAACTGCAACTGCTACAGCGA-------------------------------------------------C------------------AGCGACAGCCACAGCAACAAAAAGCTCAAGCAAATAA-------ATGAAGATA--------A------AAACAATAACAACAAGTA---------------------------------A---AAAAAAGTGCGGCTGCAGCTGAGAGTCGCTTGTAATTACCGCAACGATCAACGAACACCGATCAACGTCG---AC----------------------- | |
| droWil2 | scf2_1100000004902:11023529-11023740 + | GACGC-A-----GCCCGGTAC----CG---CTG---------------TTG------CTG----------------------------TTGCCATTGAAGG-----------------AT---------------------------------------------------------------------------------------------------------GTG----------------------------------------------CG-CTTTGGCTTTTGAAAAGA-----------------------------TTTCGATTTTTGGGCTCT--------------------------------TTTCAATTG-----------------------TTCG-------------------CACG-------G-------------TAGCAACGGCAGC---A---GCAACAACAACAACAACAACAGCAA----------------------------------CAGC---AA------C------------------G---------------GCGG------------------------------------------AGCCCA-----------------------------------------AAAC------------TAATTTGCGGCTAACGTTTTAGGTTGCCGGAAATGGCTAAGGCCATTAGA----------------------G----------------------- | |
| dp5 | XR_group6:12594695-12594892 + | AACTC-A-----G-CAACAGCA-GCAA---CAC---------------TCA------CAT----------------------------CAG------------------CAGCAACAGTTGCAACAG------------------------------------------------------CACCC---------CCAGCAGCAGCAGC---------------------------AGCAGCA-----------ACAGCAACAGC--------------------------A------------------------------------------A--------------------------------------------------------------------------------------TCTTTTGGCTTAGCCGACAC-C------------AGCAACAACAAC---A---ACAACAACAACAGCAGCAACAACAG-------------------------------------------------------------------TGGC---------------------------------------------------------AA------CAGCAG--------------CAGCA-ACAGCAA---------------------------------------------------------------------------------------------CAACGGTGTCTCCTCGAAGTCGTTTGT | |
| droPer2 | scaffold_58:329285-329478 - | ACCTC-A-----CGCAGGACA-CGCAG---GGC---ACGCA-GGAAGGAGGCGGTGGCAG----------------------------TGG------------------CGGTGGCGGTGGCATAC---------------------------------------------------------------------GAT--------A--GAAGCATT-------CGTG-----------------------------------------------------TGCTTT-----------------------TAGCTTGAATACCAAAACAATTTGAGCGCACAGTAAAATTCAATGGACGCAGGAGTAAA-----------------------------------------------------------------------------------------AACGGAA---------------CC----------------------------------------------------------AATAGA---------------------------------------GAAAG-----AGA------------------------------A------AATCAGCAGA--------------------------AGCAAA--------AAAAA-------------------------------------------------------------------------AA----------------------- | |
| droAna3 | scaffold_13047:1344818-1344951 + | ACTGC-A-----AC-TGCAAC-------------------------------------TGCAACTGGCAATTGATTTGGTC-------------------------------------------------------AACATACGGCCTGCTGATTGCCACTGACTTGGTT----------------------------------------T-----C-------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTG----AC---TAA-----------------------------------------------TGA-------------TACAAACGACAAC---AGCAACTGCAACACTAGCAACAACAACAC-------------------------------------------------------------------TAGC---------------------------------------------------------A----------------------------------------------------------------------------------------------------------------------------------------A----------------------- | |
| droBip1 | scf7180000396641:1028401-1028551 + | ATTCC-C-----TGCACCAGCA-GCAA---CAG---------------TCG------CAA----------------------------CAG------------------CAGTCGCAGCAGCAGCAG------------------------------------------------------CAGCT---------CAA--------------------------------------------------------CCACCAGCAGC--------------------------T------------------------------------------AACGCACC-------------------------------------------------------AGCAGCAACAGGCGCT-------------------------------------GC------ACCAGCAAC---T---GTCGCACCAGCAGCAGCAACAGCAGCA-------------GCAG---------CAA---CAAC---AC------C------------------C-------------------------------------------------------------------------------------------------------------------------G-------------------------------------------------------------------------CA----------------------- | |
| droKik1 | scf7180000302388:1127968-1128127 - | CCTA--------TGCAACTGGC-ATAT---TTG---------------TTG------CAG----------------------------TTG------------------CAGTTGCAGTTGCACAT---------------------------------------------------------------------ATTGTTACTGCA---------------------------------------------------------------------------------------------------------ATTTGCTTAACCGAT-----CA-----------------------------------C----TTTTGCTAA-------------------------------------------------------TTCGCAGCACATTCGCTGGCA------AC---A---A-----------------------------------AAGAGCTG---------CGA---CGGC---GGCGGT---------------------------------------ACAAG-----AACATG-------GCGCCAAAAAC-------------------------------------------------------------------AAGA-------------------------------------------------------------------------AA----------------------- | |
| droFic1 | scf7180000453872:482986-483199 + | CAATGCAAACCAA------GCAAGCAA---CAA---------------CAA------CCA-------------------------------CCATGAGAGCAACAGCAACA---CCACCGCCACCT---------------------------------------TGGTGGTCAACACCAGCAGCA---------CT-TCAGCAGCAGC---------------------------AGCAACATCAGCAGCAGG--------------------------------------------------------------------------------------------------------------------AGCA---------AC---ATCAGCAGCA-------------------------------------------------------------------ACTTCG---AC------------------------------C-TCGGCAGCAG---------CAGCAA---CTGCAACTGCAACGGCAAGTGCAACAGCAGCAGC---------------------------------------------------------AA------CATCAG----------CCA----------------------------------------------------------------------------------------------CGAACAACAACC---AC----------------------- | |
| droEle1 | scf7180000491212:1887309-1887454 - | CCAC--A-----AGCAGATGCA-GTTA---CGG---------------ATG------CAG----------------------------TTG------------------CAGTTGCGGTTGCAGTGG------------------------------------------------------CATCC---------GTG--------------------------------------------------------CCTA--------------------------ATTTAATCAAA------------T--TAGCTGAGAAATG--------------------------------------------------------------------------------------CAATGGAATGAATTTGTTCGTTTGGTCAGAGA-------------CAGAAACGGCAAC---AGAAACAGAAG-------------------------------------------------------------------C------------------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GA----------------------- | |
| droRho1 | scf7180000779337:100077-100252 - | CCCTC-T-----TTCATCCGC---------CGG---------------TTG------TGG----------TCGCTTTTAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAC------------TCATTGCCTCGTAATTAGTGGAAATTACCTGCGCAGTCCCCTGCTCTATCTCCCAAAAAAAA--------------------------------------------------AATACGCTCGAAAGAGACGGGA------AATACACT-----------G---AGAAAGAGATG-------------------------------------------------------------------G--------------------------------------------------------------------------------------ACAGA---------CTCGCGGAGAT-TCGCCGC---------------------------------------------------TTGTAATT------------------------------------AC----------------------- | |
| droBia1 | scf7180000302041:838333-838508 - | ATTGC-T-----G-CTGGCCC-------------------------------------GGCAA--------CATGTTGAC----------------------------------------------------------------------------------------------------------AAGGTCAGC--------------GT-----CTAATGGAAATTGACGGCGGTGCAA-------------CCGCA---------------GTCATCTATCTCTTCCTCTTC------------CTCTCGCCAAGTG-TGAGTGCATAAAT-----------------------------------C----ATGCGTTGC------------------------------------------------------------CCAGACGAC------AACAGCAGC---A---ACAGCAACAGCAACAGCAACAGCAA-------------------------------------------------C------------------AGC---------------------------------------------------------A----------------------------------------------------------------------------------------------------------------------------------------A----------------------- | |
| droTak1 | scf7180000415795:343236-343380 + | CCTC--A-----TGCATGCGC--CCAA---CTA---------------TC---------------------------------------------------------------------------------------------------------------------------------------------------------------CT-----TCAATGTGAGTG------------------------------------------------GATTTTTCCTTTGCACAGATCATATAACTAATTCCTATCTACCAAACAGTCAAAATCT------------------------------------CACG---------AC---CAATGCCACG-------------------------------------------------------------------CCC---------------------------------ACCAGC-GCA-CAGCTGCAACAGCAGCAG---CA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC----------------------- | |
| droEug1 | scf7180000409471:1596663-1596851 + | AGCAG-G-----TC-AAGAGCCAACAG---CTG---------------AAG------CAG----------------------------CAA------------------CATTCGGCATTGGTCAA---------------------------------------------------GTTGCT---GGAATCGGC--------------GC-----CCATTAACA-------------------------------AGCAACAG--------------------------------------------------------------------CAGACGCCCA-------------------------------------------------------AACAAATTGTTTACCT-------------------------------------GC------AACAGCAAC---A---ACAGCAGCAGCAGCAGCAACAACACCAGCAGCC-A------------------------CAGC---AA------C------------------C---------------GCAA------------------------------------------CGTAAA-----------------------------------------AGACTGAAAAACGA-------------------------------------------------------------------------AG----------------------- | |
| dm3 | chrX:5446238-5446430 + | ACATG-C--------AA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAGC---------------------------AGCAGCAGCAGCAGTGGA--------------------------------------------------------------------------------------------------------------------AGCA---------ACATCAATTTCAACA-------------------------------------------------------------------TCA---------------------------------TTCAACTTCAGGAGCAG---------CAA---CAAC---AGCGAC---------------------------------------AAAAG-----GGCAATTAAGTAGGCC--CAAGATCT--CGGCAA------GAAAG---------AGTTAAGAGCA-CCCAAGA----------------------------------------------AGGGCCTAGAAT----GAAACAATCATAGGATGTC--------------------TTACATTTAAGCCAAGACC | |
| droSim2 | 3r:20059949-20060148 + | GGCCC-A-----GGCAC-AGGA-GCAA---CAC---------------TTG------CAG----------------------------CAG---------------CAACA---CCAGCAGCACCA---------------------------------------------------GTTGCAGCT---------CTTGCAACTGCAGC---------------------------AGCAGCAACAGCAGTTGC--------------------------------------------------------------------------------------------------------------------AGCA---------AC---CAGTGCAGCA-------------------------------------------------------------------CCA---------------------------------TCCGCC-GCAGCAGCTG---------CAA---CAGC---AGCAGCAGCAGCAGGTG---------------------------------------------------GCT--CAGGATCCAGCAGCAA------GTGCA---------TTCGAGGAGCA-CCAGCGA---------------------------------------------------------TTGGT------------------------------GCAGC----------------------- | |
| droSec2 | scaffold_0:20880743-20880945 + | GGCCC-A-----GGCAC-AGGA-GCAA---CAC---------------TTG------CAG----------------------------CAG---------------CAACA---CCAGCAGCACCA---------------------------------------------------GTTGCAGCT---------CTTGCAACTGCAGC---------------------------AGCAGCAACAGCAGTTGC--------------------------------------------------------------------------------------------------------------------AGCA---------AC---CAGTGCAGCA-------------------------------------------------------------------CCA---------------------------------TCCGCC-GCAGCAGCTG---------CAACAA---CAGCAGCAGCAGCAGCAGGTG---------------------------------------------------GCT--CAGGATCCAGCAGCAA------GTGCA---------TTCGAGGAGCA-CCAGCGA---------------------------------------------------------TTGGT------------------------------GCAGC----------------------- | |
| droYak3 | 3L:7523671-7523913 + | AGCAG-C-----A----------GCAACAACTAATCCTGCTG------CCC------CAG---TTGGATCCTAATGTTGTCAACACGT---CCACGGCAGCAGCAGCGGCA---C-----------------------------------------------AGCTCGTC--CA---GTTGCAGCA---------GCAGCATCAACAGC---------------------------AGCAGCAGCTGCAGCAAC--------------------------------------------------------------------------------------------------------------------AGCA---------AC---TGCAGCAGCA-------------------------------------------------------------------ACAGCA---GC------------------------------T-GCAGCAGCAA---------CAGCAA---CTGCAACAGCAACAGCAGCTGCAAC----------------------------------------TC-----AAT--CAGCAG-CAGCAGCAA------CTC-----------TACCAGCAGCA-ACAGCAA---------------------------------------------------------TA------------------------------CCT---GC----------------------- | |
| droEre2 | scaffold_4784:11285393-11285587 - | AGCAG-C--------AGCAGCA-GCAA---GCG---------------TTG------CAT----------------------------CTG------------------CAGCAGCAACAGCAACAACATTCCAGTAACATGGCACTAAAACGTGATCGTGATCGTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTCTCAACGTCAAA-------------TGGCAACGGCAAC---AGCAACAGCAACAGCAACAGCAGCAACAC-------------------------------------------------------------------CAAC------A------ACAA----------CACCTC-------A--CTGGACCCAACAGCAA-----------------------------CATCCAGCAA---------------------------------------------------------C---T------------------------------GCAGC----------------------T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:42 PM