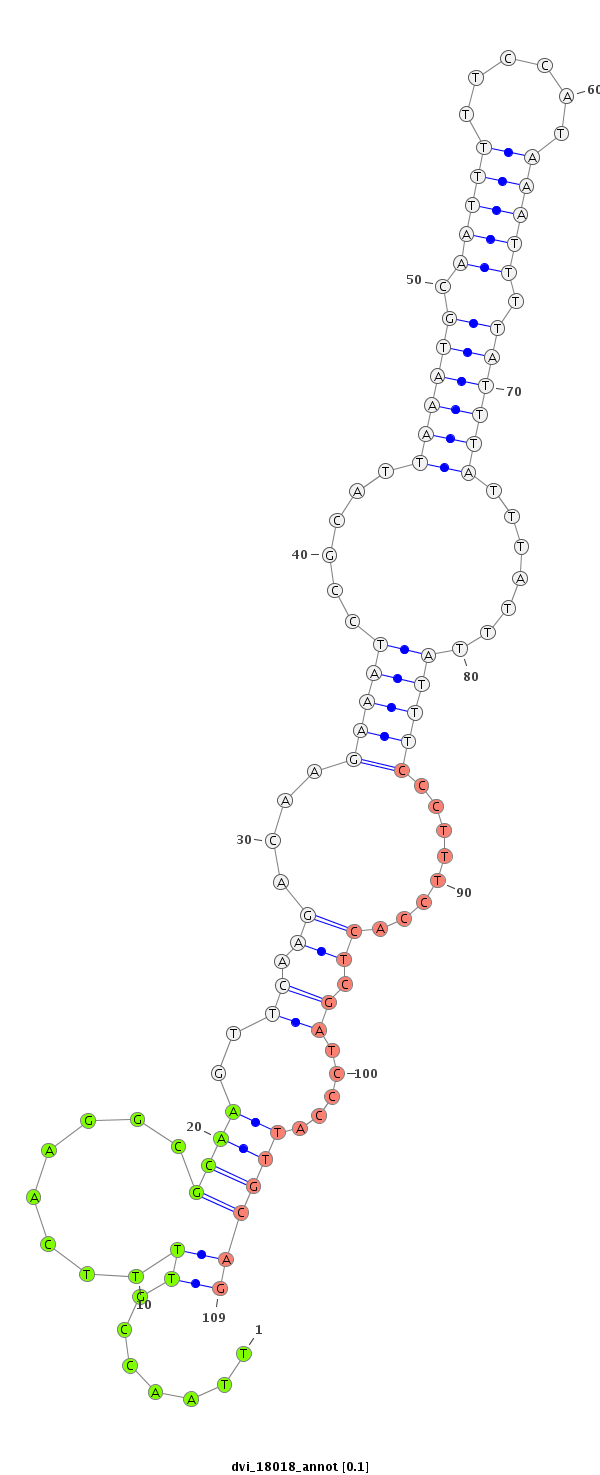
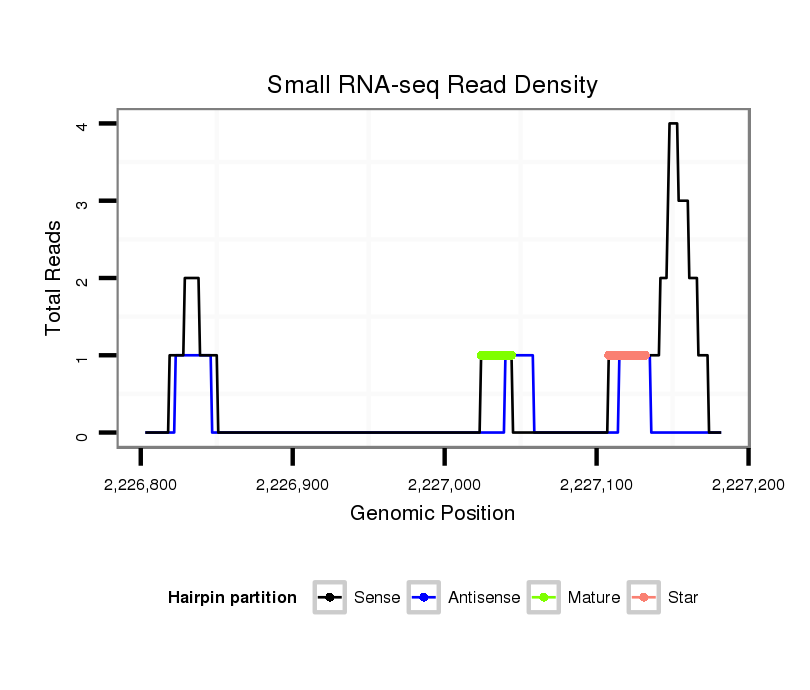
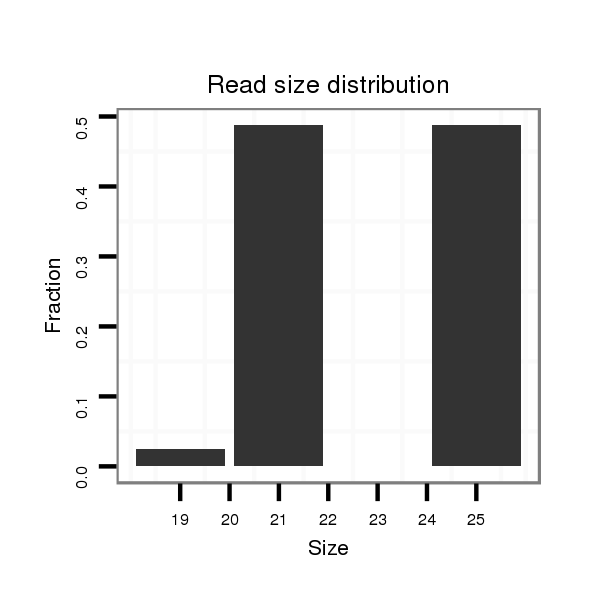
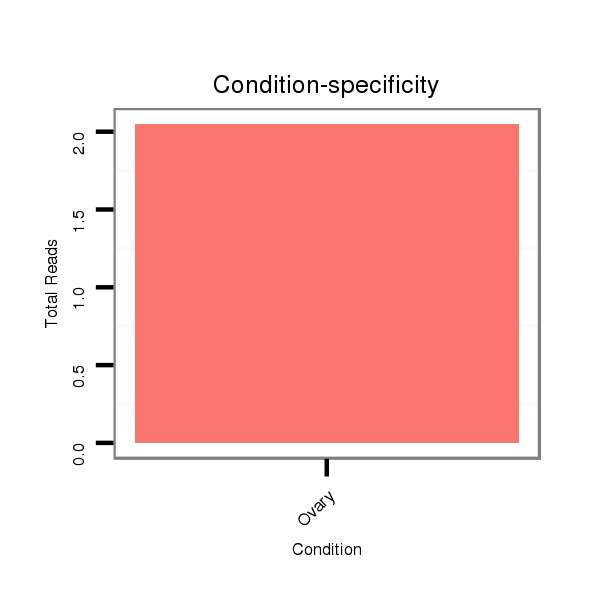
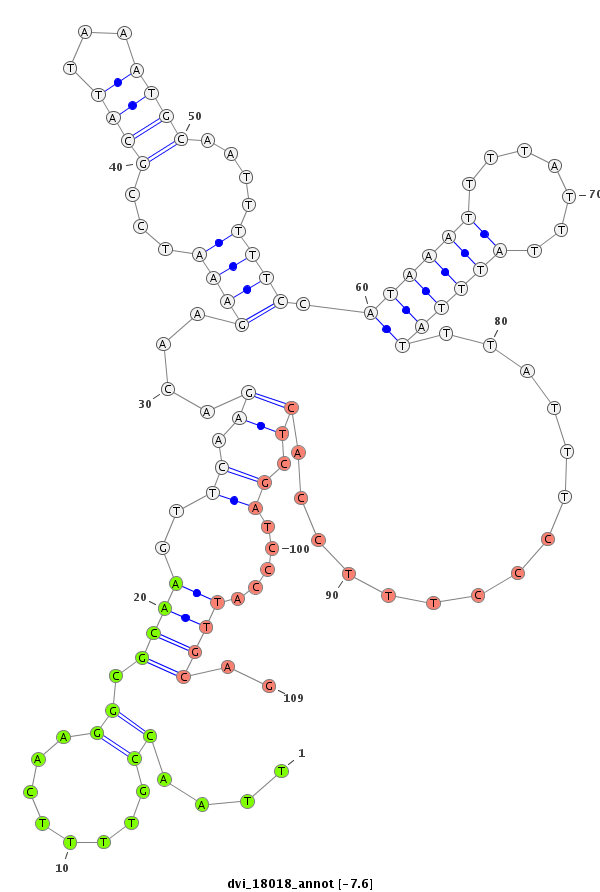
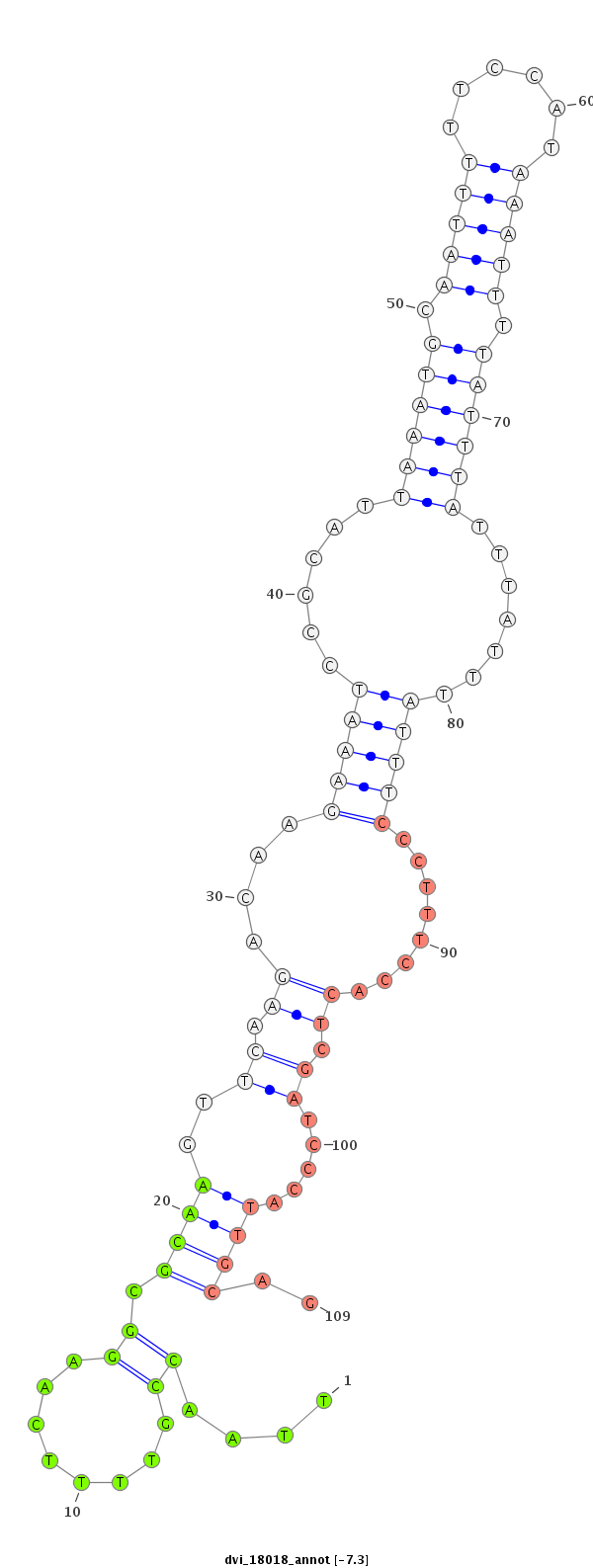
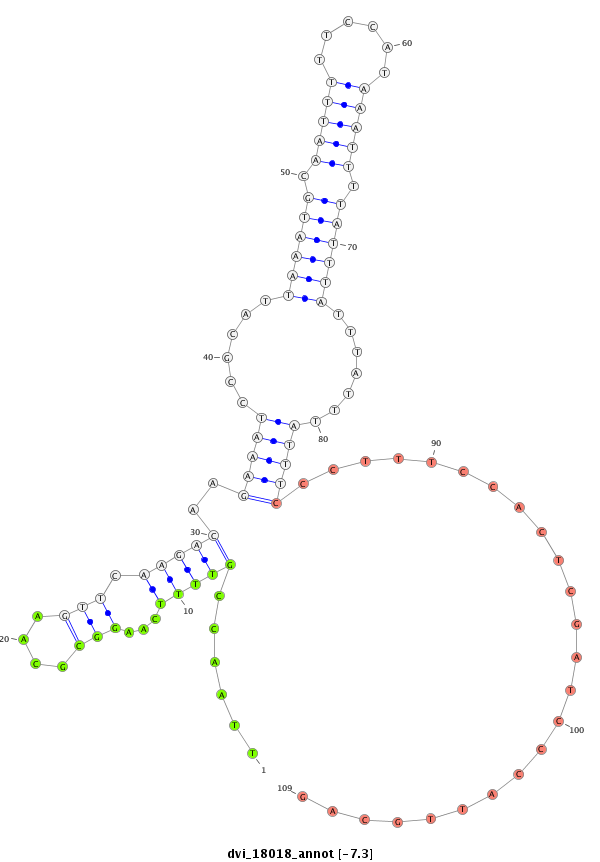
ID:dvi_18018 |
Coordinate:scaffold_13049:2226853-2227132 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
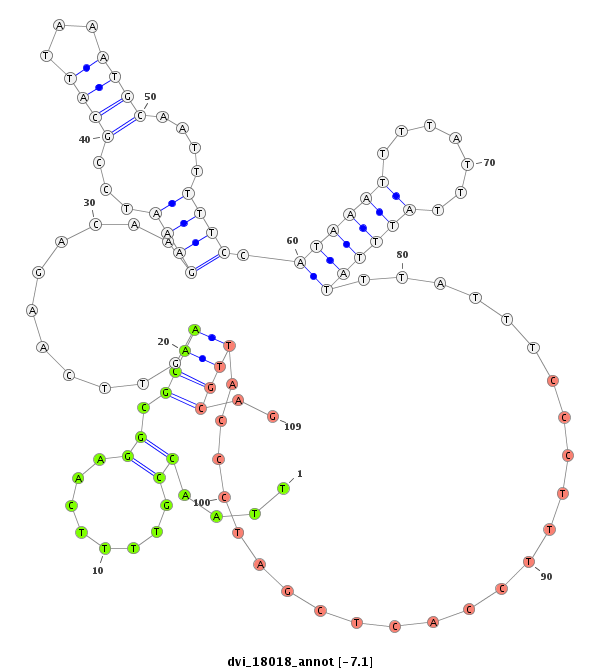
| -7.6 | -7.3 | -7.3 | -7.1 |
 |
 |
 |
 |
CDS [Dvir\GJ12854-cds]; CDS [Dvir\GJ12854-cds]; exon [dvir_GLEANR_12795:2]; exon [dvir_GLEANR_12795:3]; intron [Dvir\GJ12854-in]
| Name | Class | Family | Strand |
| A-rich | Low_complexity | Low_complexity | + |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GGAGCGGCGCTCCATAGACTACGGCGATCATCATGCGAATAAAGTGTCGCGTAAGTAAATTTAAATGCATTTAAATAAGGTCACAAAGTCAGTCAACCCATGGCAACGGTATCTGAATCCGAATCGGAATCGGAACTGCAAAACACGTTTACCACAAAAAAAAAAAAAAATCAATACATTAATTAAACAGAAAAAAAAAAAGAAAAAACCAAAAAAACAAATTAACCGTTTTCAAGGCGCAAGTTCAAGACAAGAAATCCGCATTAAATGCAATTTTTCCATAAATTTTATTTATTTATTTATTTCCCTTTCCACTCGATCCCATTGCAGATGAGAAAATGCACAAGGGTCAGCATTATGTCGGACGCAGCAGCTATCCG *****************************************************************************************************************************************************************************************************************************.......((........((((..((.((....(((((......((((((.(((((......))))).)))))).......)))))........)).)).....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060666 160_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060656 9x160_ovaries_total |
SRR060658 140_ovaries_total |
SRR060673 9_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR060678 9x140_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR1106727 larvae |
V053 head |
SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060665 9_females_carcasses_total |
M028 head |
SRR060674 9x140_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TTAACCGTTTTCAAGGCGCAA.......................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GACTACGGCGATCATCATGC........................................................................................................................................................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CCCTTTCCACTCGATCCCATTGCAG.................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GAATAAAGTGTCGCATGAGAAA.................................................................................................................................................................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........CCATAGACTACGGCGATCATA............................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................AGGGTCAGCATTATGTCGGACGCAGC......... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................AAGGGTCAGCATTATGTCGG................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TACATTAGTTAAACTGAAAAAAAAAAAA.................................................................................................................................................................................. | 28 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................TTTCCCTTTCGACACGTTCCCA........................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................TGCACAAGGGTCAGCATTA...................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................ATGAGAAAATGCACAAGGGTC............................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................ATCATCATGCGAATAAAGTGTC............................................................................................................................................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGCAACGGTATCTGAAT...................................................................................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................AAAAAAAAAAGGAAAAACCAAAAAA.................................................................................................................................................................... | 25 | 1 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................GCAACGGACTCTTAATCCG................................................................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.15 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ACTTTAATGGAACAGAAAAAAAA...................................................................................................................................................................................... | 23 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGCAACGGTATCTGAAT...................................................................................................................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................AATCCCAATCGGAAACTGAAC.................................................................................................................................................................................................................................................... | 21 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................GAGAAAATGCACAAAGGACG............................ | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ACAGGGTGACCACAAAAAAAAA........................................................................................................................................................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................TTAAACGGAAAAAAAAAA.................................................................................................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................ACCACAAAAAAAAAAAAAA................................................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................TCCCATTGTAGAGGTGAAA.......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CCTCGCCGCGAGGTATCTGATGCCGCTAGTAGTACGCTTATTTCACAGCGCATTCATTTAAATTTACGTAAATTTATTCCAGTGTTTCAGTCAGTTGGGTACCGTTGCCATAGACTTAGGCTTAGCCTTAGCCTTGACGTTTTGTGCAAATGGTGTTTTTTTTTTTTTTTAGTTATGTAATTAATTTGTCTTTTTTTTTTTCTTTTTTGGTTTTTTTGTTTAATTGGCAAAAGTTCCGCGTTCAAGTTCTGTTCTTTAGGCGTAATTTACGTTAAAAAGGTATTTAAAATAAATAAATAAATAAAGGGAAAGGTGAGCTAGGGTAACGTCTACTCTTTTACGTGTTCCCAGTCGTAATACAGCCTGCGTCGTCGATAGGC
**************************************************.......((........((((..((.((....(((((......((((((.(((((......))))).)))))).......)))))........)).)).....))))))***************************************************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
V047 embryo |
SRR060658 140_ovaries_total |
SRR060673 9_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
SRR060676 9xArg_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060657 140_testes_total |
M061 embryo |
SRR060681 Argx9_testes_total |
GSM1528803 follicle cells |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................CAGCCATGACGTTTAGTGCAA....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CGTTGCCATAGGCTTAGGCTT................................................................................................................................................................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CTCAGCCATGACGTTTAGTGC......................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................GTGAGCTAGGGTAACGTCTAC............................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TGCCGCTAGTAGTACGCTTATTTC................................................................................................................................................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................GCGTTCAAGTTCTGTTCTT............................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TCAGCCATGACGTTTAGTGC......................................................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGAGGAAGCTGATGCCGCAAGT.............................................................................................................................................................................................................................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................GGCTTTTTTGGTTAATAGGC........................................................................................................................................................ | 20 | 3 | 10 | 0.50 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........AGGTATCTGATGCCGAAAG............................................................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CTCAGCCATGACGTTTAGTG.......................................................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................GTCTGGTGGGTTCCGTTGCC............................................................................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AGCCATGACGTTTAGTGCAAG...................................................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TTCCGTTAAAAAGGTATTTAA............................................................................................. | 21 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GCCATGACGTTTAGTGCAAG...................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TTCCGTTAAAAAGGTACTTAA............................................................................................. | 21 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GCAGTAGACTGAGGCTTAGC.............................................................................................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................AGCGTACAACTTCTGTTCTT............................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................GGGGATTTAAAAGAAATAAAT.................................................................................. | 21 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TTACGTTAAAAAGGTATTTTA............................................................................................. | 21 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TTACGTTAAAAAGGTATTT............................................................................................... | 19 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................AGACTTAGGCTTAGACTTA.......................................................................................................................................................................................................................................................... | 19 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CCATGACGTTTAGTGCAAG...................................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................GTCGGGTAGCGTTGCGATA............................................................................................................................................................................................................................................................................ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................TCGTGCGACAGCCTGCGTC.......... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGTGTTTTTTTTTTTTTT................................................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................TCTTTTTTTTTTTCTTTT.............................................................................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:2226803-2227182 + | dvi_18018 | GGA----GCGGCGCTCCATAGACTACGGCGATCATCATGCGAATAAAGTGTCGCGTAAGTAAATTT--------------------------AA--------------------------------------AT-----------------------------------------------------------------------------GCATTTAAATAAGGTCACAA--AGTCAGTCAACCCA--TGG-C--AACGGTATCTGAATCCGAATCGGAATCGGAACTGC------------AAAACACGTTTACC---ACAAAAAAAA----AA-A--AAAATC------AATACATTAATT-AAA-----CAGA--------------------------------------AAAAAAA----AAAGAAAAAACCAAAAAAA------CAAA-TTAACCGTTTTCAAGGC--GCAAGTTCAAGACAAGAAATC----C--GCATTAAATG-----CAATTTTTCC----ATAAATTTTATTTATTTA----T-------TTATT--TCCCTT-------TCC--ACTCGATC----CCATTGCAGATGAGAAAATGCACAAGGGT---CAGCATTATGTCGGACGCAGCAGCTATCCG |
| droMoj3 | scaffold_6654:1636772-1637137 - | AGA----GAGACGGTCTATAGACTCCAGCTGGCATCATAAAAATAAGCTTTCACGTAAGTATAAACATATATGTATATCTCAATTTAGTATGAA--------------------------------------AT----------------------------------------------------------------------------GCATATTTAATAAGGTCACAA--AGTCAGTCAACCCA--GAGTC--AAAAGGA-----------AACGAACTCGGAAATGTT------TACAAAAACTAT------------------A--------A--AAAATC------AATAAAAATATTTAAAATAT-TTAA--------------------------------------TATTTAA----ACAAAGCGC----------------AGTC-TTACACGATTTCAAGGC--GCAAGTTCAAGTTTA---AAT----TGA---AATGTAATATTT----------TTCAAAATAATTT--------ATT--T-------TTATTCT--TCTT-------------TCT--GC----CCTTTGCAGATGGCAAGATGGATAAGAAGACGCCACATGGCGTCAAGCGCAGCAGCTTTCCA | |
| droGri2 | scaffold_15110:3267054-3267380 + | GGA----GCGACGGTCCATAGACTACAGTGGTCATCACGAAAATAAACTAGCACGTAAGTGTATTC--------------------------AG--------------------------------------CG-----------------------------------------------------------------------------ACGCTTAAATAAGGCCACAG--AGTCAGTCAACCCG--AT-----G--TGCATT-----------------GAGAAACGGA-------ATCGGAAACACAAA----------CACACAAACAATT-C--ATAATT------AATAAT---------------TCAA------------------------------------------TAA----ATAATGCAGTTCCGATTTT------AAAGCTTGCCCGTTTTCAAGGCAAGCAAGTTCAAGTGGA---GTC----T--TTATTAAATA-----CTAT--------------------------------T---TTTTT------T--GTATATGATTTTT--ATGC--GC----TCTTTGCAGATGACAAAATGCATAAGGAT---CAGCATTATATTGGCAGGAGTAACTATCCG | |
| droWil2 | scf2_1100000004768:225797-226133 - | GGG----CAAAAGATCCGTTGACTACAAGCCGCATCATCTGAATCGTTTGGGATGTAAGTACATA----------------------------A--------------------------------------AT-----------------------------------------------------------------------------TCATAGTAATAAGGTCACCA--AGTCACTCAACTCG----------------TG-----------------CGGAACTGCG------TTAAGTA--------------------------------A--ATAATT------AATAAATTGATA-ATA-----ACAA-AACAAAAACGAAAAGAAAAAAAAAAATCACAAAATCACATTTAC----ATAAAACAGTTCCGTTTTTCGCATTCAAC-TTAACCGTTTTCAAGGC--TCCAGTTCAAGTTCG---AAT----TGT--AATTGTAA-----------------------------------------T-----------TGT--TGTTTTTGT---------GC--T-----TTTCCACAGTTGGCAAACTGCACGACAAA---CATCATTACATCAGTCGCGGCAACTTTCCA | |
| dp5 | XR_group8:687893-688225 + | GGA----CAAGCGGTCTATAGAGTACAGCCATCACCATCTGAATCGCCTGGGATGTAAGTATAAG---------------------------------------------------------------------------------------------------------------------------------TCAGATAAATCAGATTATTGACTGAATAAGGTCAGAG--AGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGG----------CAAAAACACGTTT------AAATTTGTAG----AC-G--GCC-----------TGTCGGCAAT-GCT-----CGGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAC-TTAACCGTTTTCAAGGC--ACAAGTTCAAGATCA---CTT----TTCTCAC--ACAA-----------------------------------------T-----------------GTTTCCCATTTTT--GTTC--AC----TGTTCACAGACGGCAAACTGCACGACAAG---CATCATTACATCACCCGGAGTAACTATCCC | |
| droPer2 | scaffold_32:796052-796384 - | GGA----CAAGCGGTCTATAGAGTACAGCCATCACCATCTGAATCGCCTGGGATGTAAGTATAAG---------------------------------------------------------------------------------------------------------------------------------TCAGATAAATCAGATTATTGACTGAATAAGGTCAGAG--AGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGG----------CAAAAACACGTTT------AAATTTGTAG----AC-G--GCC-----------TGTCGGCAAT-GCT-----CGGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAC-TTAACCGTTTTCAAGGC--ACAAGTTCAAGATCA---CTT----TTCTCAC--ACAA-----------------------------------------T-----------------GTTTCCCATTTTT--GTTC--AC----TGTTCACAGACGGCAAACTGCACGACAAG---CATCATTACATCACCCGGAGTAACTATCCC | |
| droAna3 | scaffold_13337:7154473-7154865 - | GGAGGGACAGACGATCCATCGACTATCCACACCATCACATGAATCGGCTAGGATGTAAGTATCTTC--------------------------CACC-----------------------------------------------------------------GATTAGTCTC------TGTA-----------------------AAGATTAGCAATTGATTGAGGTCAGAG--TGTCAGTCAACCCG--AC-AGAGAATTGCATT-----------------CGGAACTGAAATGCAGCACCGAAAACACGTTCCC-----AGCGTGTAG----ACAA--GTAATT------AATAAACACATT-ACATTACTCGGGGAAT----------------------------------AATTTGC----ATAAAGCAGTTCCGATGCA------ACAG-TTAACCGTTTTCAAGGC--GCAGGTTCAAGATCA---ATCGATTAGC----TGGCT----------------CTCAA-----------------CTGACGTTTTCTT------TCCGTTTCCCCCTTTTT-T-----------TTTGTCCAGATGGCAAGATTCACGACAAA---CATCATTACATCAGCCGCAGCAACTATCCG | |
| droBip1 | scf7180000396418:162563-162990 - | GGAGAGATAGGCGATCCATCGACTATCCACACCATCACTTGAATCGGCTGGGATGTAAGTATCTTC--------------------------TATCTT----------------------------TTGATAGCACTCAA-GG-GAAATTCAAAAAATTTAAATTAGTCTGT-GATCT-TA---------ATTG------GAATAAGATTAGTAATTGATTGAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGAAATGCAGCACCGAAAACACGTTCCC-----AGCGTGTAG----ACAA--GTAATT------AATAAACACATT-ACT-----CGGGCCAT----------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------ACAG-TTAACCGTTTTCAAGGC--ACAGGTTCAAGATCA---ATCGATTAGC----TGGCT----------------CTCAA-----------------CTCATGTTTTCTT------TCCGTTTCC---------------AACTTTTTCCAACAGATGGCAAGCTTCACGACAAA---CATCATTACATCAGCCGGAGTATCTATCCG | |
| droKik1 | scf7180000302486:875916-876276 - | GGA----CAGGCGGTCCATTGAGTACTCGCACCATCATCTAAATCGGCTGGGATGTAAGTATCTTG--------------------------AA--------------------------------------GAACCCAAACA-------------------------------------------------------------GAAACTAAAGAGAGATTAAGGCCAGAGGGTGTCAGTCAACTCCAGAC-AGATAATTGCATT-----------------CGGAACTGCAGT----GGCCGAAAACACGTTTT-A---AAATGTGTAG----AC-TTCGGAATTAAAATCAATATT---------------------------------------------------------CATTTGCATACACAAAGCAGTTCCGATGCA------ACAG-TTAACCGACTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTCGCTC------------------------------------A----C-----------------GTTTCCCCCTTTCT-GGTT--TT----CCTTTCTAGACGGCAAAGCGTACGACAAG---CATCACTACCTCATCCGGAGCAGCTTCCCG | |
| droFic1 | scf7180000454107:497103-497371 - | G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGATCGAATAAGGTCAGAG--TGTCAGTCAACTCG--AC-----AATTGCACT-----------------CGGAACTGCA------TATCGAAAACACGTTTT-A---AAGTGTTTAG----GT-T--GCAATT------AATACGCCTTTA-AGT-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------ACAG-TTAACCGATTTCAAGGC--GCAGGTTCAAGATCG---ATT----TGCTCACTCGCAT-----------------------------------------C-----------------GTTTCCCCCTTTTT-TTCC--AA----CTTTTTCAGACGGCAAAACACACAACAAA---CATCACCATATCAGCCGGAGCAGTTATCCA | |
| droEle1 | scf7180000491255:3462902-3463329 - | CGA----CAGGCGATCCATCGAGTACCCGCATCACCATCTAAACCGACTGGGATGTAAGTATCTAACTAGAT-----------------------CTTTAAGCTTATTAAGCTATTAATATTTGATTTGAT------------TAAAACTTGTAAAAGTTA---TAGTCTGTAGATCTGTAAGGAAAAGAACAGGC-----AATTAAATTAGCAACTAAATAAGGTCAGAG--TGTCAGTCAACCTG--AC-----AATTGCATT-----------------CGGAACTGCA------CACAGAAAACACGTTTTTG---AAGTGTGTAG----AT-T--GGAATT------AATGTTCG-------------------------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------ACAG-TTAACCGATTTCAAGGC--GCAGGTTCAAGATCG---ATT----TGCTCACTCGCTC------------------------------------ACT--C-----------------GTTTCCCCCCTTTTCCCCT--GAC---TTTTTTCAGACGGCAAAACGCACAACAAG---CACCATTCAATCAGCCGGAGCACTTACCCA | |
| droRho1 | scf7180000779902:37215-37482 + | AAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATAATTAGCAACTAAATAAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TACAGAAAACACGTTTTTA---AAGTGTGTAG----AT-T--GGAATT------AATAAGCG-------------------------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------ACAG-TTAACCGACTTCAAGGC--GCAGGTTCAAGATCG---ATT----TACTCACTCGCAA-----------------------------------------C-----------------GTTTCCCCCTTTT---TCC--GA----CCTTTTCAGACGGCAAAACACACAACAAG---CACCATCATATAAGACGAAGCAGTTATCCG | |
| droBia1 | scf7180000302428:3086331-3086589 + | G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGACTGAATAAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------AATCGAAAACACGTTTTTA---AAGTGTGTAG----A--T--GCAATT------AATGCGCGCATT-ACA-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGATCTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTCGCAA-----------------------------------------C-----------------GCTTTCCC-------------GA----CTATTTCAGACGGCAAAACACACAACAAG---AACCATCACATCCGGCGGAGCAGCTATCCC | |
| droTak1 | scf7180000415352:331862-332139 + | TAA----GGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAGAAAATTAGCGACTTAATAAGGTCAGAG--TGTCAGTCAACCCT--AC-----AATTGCATT-----------------CGGAACTGCA------TTTCGAAAACACGTTTATA---AAGTGTGTAG----AT-T--GCAATTA-----AATACGCG-------------------------------------------------------GATTTAC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGACTTCAAGGC--GCAGGTTCAAGATCA---ATT----GGCTCACTCGCAA-----------------------------------------C-----------------GTTTCCCCTTTTTTATTGC--GA----CTATTTCAGACGGTAAAACGCACAACAAG---CACCACCACATCAGCCGCAGTAGCTATCCA | |
| droEug1 | scf7180000409711:312011-312274 + | AT---------------------------------------------------------------------------------------------------------------------------------------------TAAA----------------------------------------------------------------ACAAATTAATAAGGTCAGAA--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAAATGCA------TATCGAAAACACGTTTTTA--AAAATGTGTAG----AT-T--GCAATT------AATACGCGCATT-ACA-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGATTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTCGCAA-----------------------------------------C-----------------GTTTCCCC--------------------TTTTTCAGATGGCAAAACGCACAACAAG---CACCACTATATCAGTCGGAGCAGTTATCCA | |
| dm3 | chr3L:17023335-17023576 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATAAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TATCGAAAACACGTTTTTTTTTAAGTGTGTAG----AT-T--GCAATT------AATAC------------------GG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGACTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTGGCAA-----------------------------------------C-----------------GTTTTCCC-------------------CCTTTTCAGCCGGCAAAACGCACAACAAG---CATCACTACATCAGCCGAAGCAGCTATCCA | |
| droSim2 | 3l:16606496-16606749 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATAAGGTCACAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TATCGAAAACACGTTTTTTTTAAAGTGTGTAG----AT-T--GCAATT------AATACGCGCATT-ACA-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTTACCGACTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTGGCTA-----------------------------------------C-----------------GTTTTTCC-------------------CCTTTTCAGCCGGCAAAACGCACAACAAG---CACCACTACATCAGCCGAAGCAGCTATCCA | |
| droSec2 | scaffold_0:9084029-9084281 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATAAGGTCACAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TATCGAAAACACGTTTTTTT-TAAGTGTGTAT----AT-T--GCAATT------AATACGCGCATT-ACA-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGACTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTGGCTA-----------------------------------------C-----------------GTTTTCCC-------------------CCTTTTCAGCCGGCAAAACGCACAACAAG---CACCACTACATCAGCCGAAGCAGCTATCCA | |
| droYak3 | 3L:17050057-17050331 + | GAT--------------------------------------------------------------------------------------------------------------------------------------------TAAAAT-----------------------------------------------------ATTGGATTAGCGAGTTAATAAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TATCGAAAACACGTTTTAT---AAGTGTGTAG----AT-T--GCAATT------AATACGCGCACT-ACG-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGATTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTGGCAA-----------------------------------------T-----------------GTTTTCCC--------------------CTTTTCAGCCGGCAAAACGGTCCACAAG---CACCACTATATCAGCCGGAGCAGTTATCCA | |
| droEre2 | scaffold_4784:16819373-16819648 + | GAT--------------------------------------------------------------------------------------------------------------------------------------------TAAAAT-----------------------------------------------------ATTGGATTAGCGAGTTAATAAGGTCAGAG--TGTCAGTCAACCCG--AC-----AATTGCATT-----------------CGGAACTGCA------TATCGAAAACACGTTTTTT---AAGTGTGTAG----AT-T--GCAATT------AATACGCGCACT-ACA-----CCGG--------------------------------------CATTTGC----ATAAAGCAGTTCCGATGCA------AAAG-TTAACCGATTTCAAGGC--GCAGGTTCAAGATCA---ATT----TGCTCACTGGCAA-----------------------------------------C-----------------GTTTTCCC-------------------CCTTTTCAGCCGGCAAATCTGTCCACAAG---CACCACTATATCAGCCGGAGCAGTTACCCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 08:44 PM