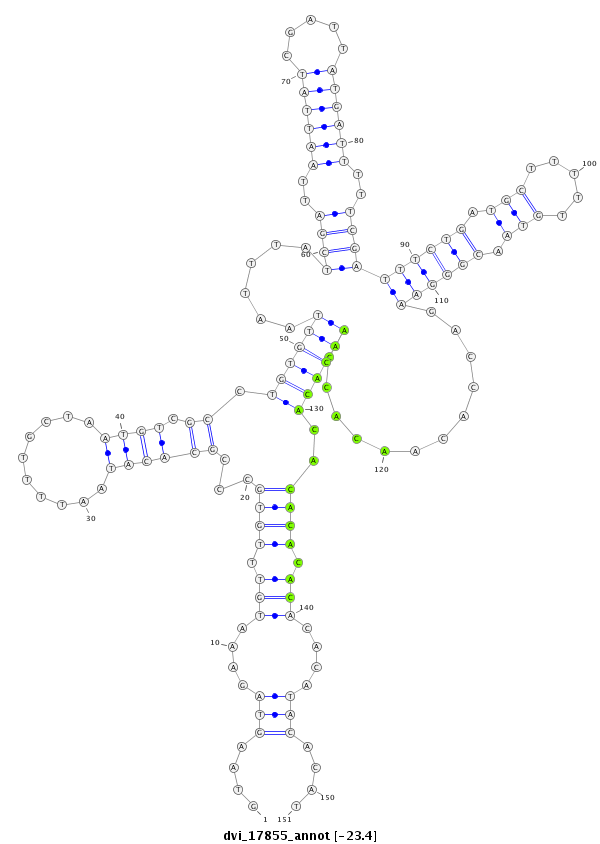
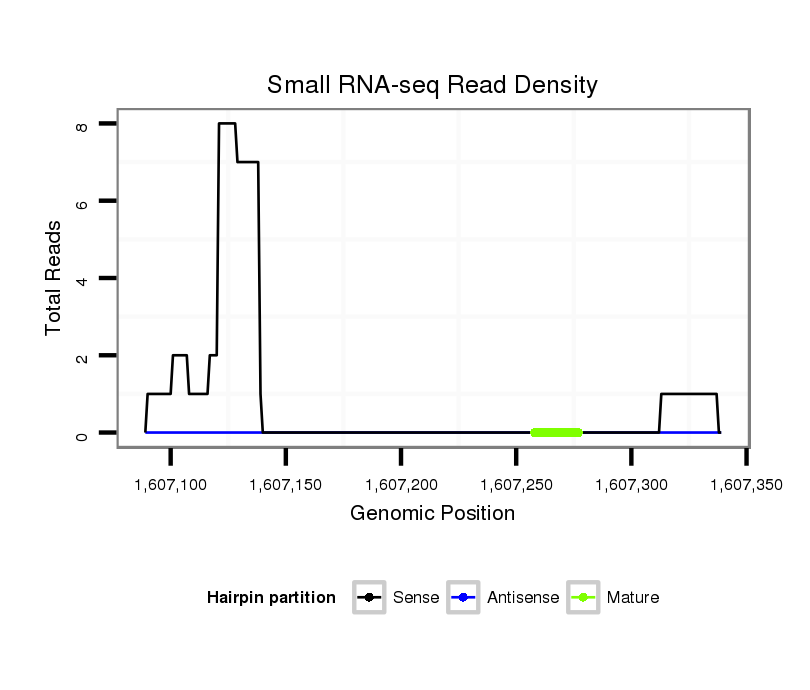
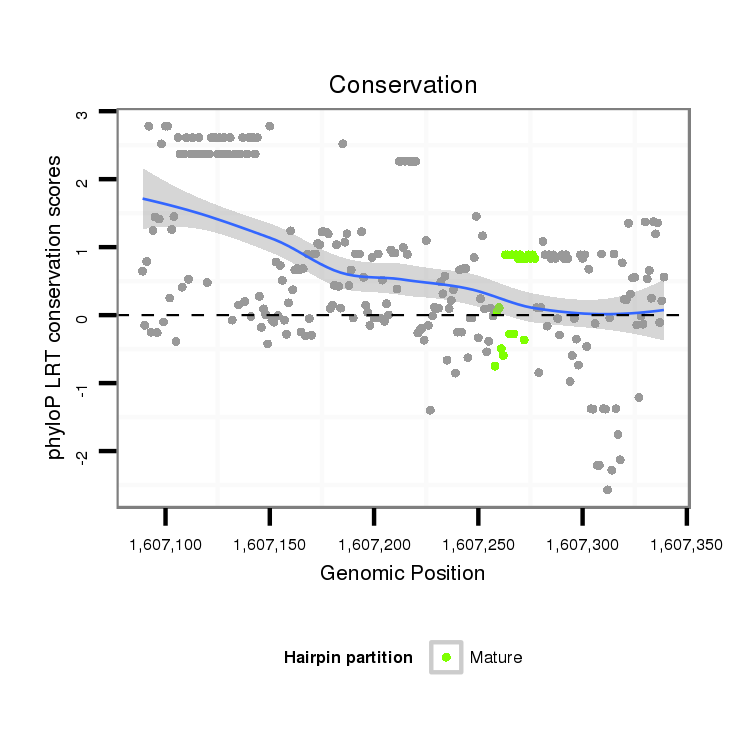
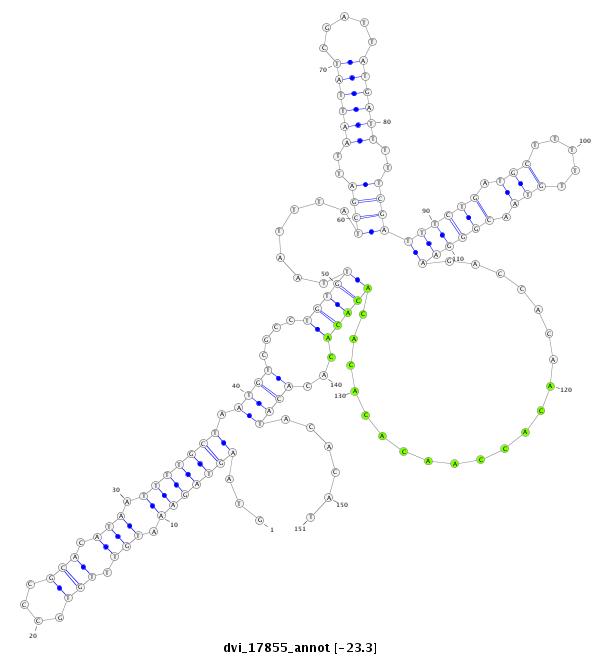
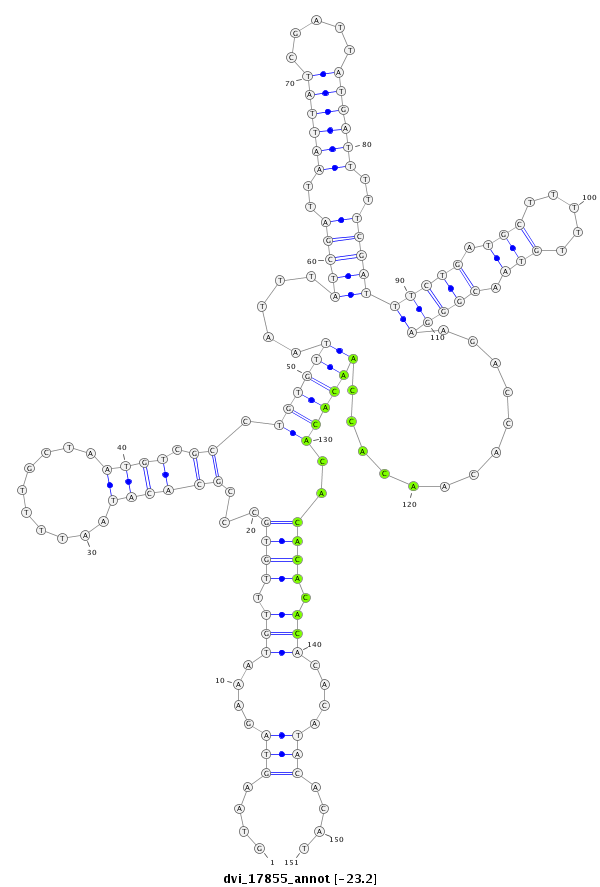
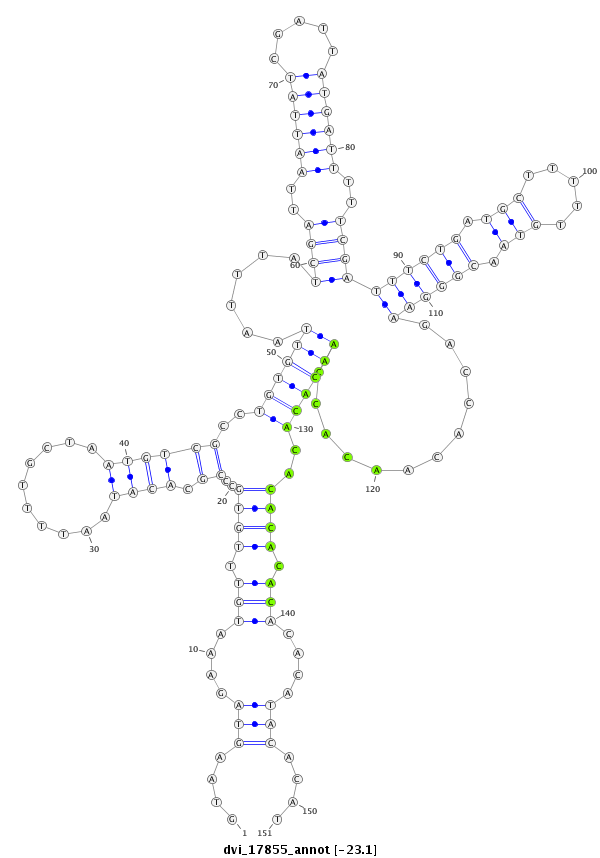
ID:dvi_17855 |
Coordinate:scaffold_13049:1607139-1607289 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.3 | -23.2 | -23.1 |
 |
 |
 |
exon [dvir_GLEANR_12748:2]; CDS [Dvir\GJ12804-cds]; intron [Dvir\GJ12804-in]
| Name | Class | Family | Strand |
| (CA)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GATTTGTTACCTTTGAGACAGAGCAGGAGGCGCAAAGACTGCAGGCGGATGTAAGTAGAAATGTTTGTGCCCGCACATAATTTTGCTAATGTCGCCTGTGTTAATTTATCGATTAATTATCGATTATGATTTTTCGATTTCTGATGCTTTTTGTAACGGGAAGACCACAACACCAACACACACACACACACACATACACATACTCACCTTACACTCACTTACAAAAACAGCTGCAAACGTTATAATGTGCC **************************************************....(((....(((.((((...((((((..........)))).)).((((((......((((..((((((.....))))))..))))((((((.(((.....))).))))))............))))))..)))).)))....)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060659 Argentina_testes_total |
M047 female body |
SRR060681 Argx9_testes_total |
M027 male body |
SRR060657 140_testes_total |
SRR060664 9_males_carcasses_total |
SRR060670 9_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
V047 embryo |
SRR060668 160x9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................CAAAGACTGCAGGCGGAT......................................................................................................................................................................................................... | 18 | 0 | 1 | 6.00 | 6 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................AAAGACTGCAGGCGGATA........................................................................................................................................................................................................ | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCAGGAGGCGCAAAGACCGCA................................................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................GCGTAAAGACTGCAGGCG............................................................................................................................................................................................................ | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................GGCGCAAAGACTGCAGGCGGATG........................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .ATTTGTTACCTTTGAGAC........................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGTAGAAATGTTTGTGCCT................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............TTGAGACAGAGCAGGAGGCGCAAAGACT................................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................AAACAGCTGCAAACGTTATAATGTG.. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GACTGAGCAGGAAGCGCAA........................................................................................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................CACACACACATACACATACTC.............................................. | 21 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................CACACACACACATACACATACT............................................... | 22 | 0 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TCACACATACACTTACTCCCC........................................... | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ACACCAACACACACACACAC.............................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CTAAACAATGGAAACTCTGTCTCGTCCTCCGCGTTTCTGACGTCCGCCTACATTCATCTTTACAAACACGGGCGTGTATTAAAACGATTACAGCGGACACAATTAAATAGCTAATTAATAGCTAATACTAAAAAGCTAAAGACTACGAAAAACATTGCCCTTCTGGTGTTGTGGTTGTGTGTGTGTGTGTGTGTATGTGTATGAGTGGAATGTGAGTGAATGTTTTTGTCGACGTTTGCAATATTACACGG
**************************************************....(((....(((.((((...((((((..........)))).)).((((((......((((..((((((.....))))))..))))((((((.(((.....))).))))))............))))))..)))).)))....)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
SRR060679 140x9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTGTGTGTGT............................................................. | 27 | 2 | 5 | 1.60 | 8 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTGTGTGT............................................................... | 25 | 2 | 6 | 1.17 | 7 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTGTGT................................................................. | 23 | 2 | 7 | 0.71 | 5 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTGT................................................................... | 21 | 2 | 15 | 0.60 | 9 | 3 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTGTGTGTGTG............................................................ | 28 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGTG.................................................................... | 20 | 2 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................CCCTTTTGGTGTTGTCGTTG.......................................................................... | 20 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CCCTGTCTCGTTCTCCACG.......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................TGGGGTTGGGGTTGTGTGT..................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................ACGTTTGGGAGATTACACG. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:1607089-1607339 + | dvi_17855 | GATTTGTTACCTTTGAGACAGAGCAGGAGGCGCAAAGACTGCAGGCGGATGTAAGTAG----------------------AAATGTTTGTGCC---C---GCACATA----------------------------ATTTTGCTAATGTCGCCTGT-----GTTA-AT--TTATCGAT--TAATTATCG-------------------------------------------------------------------ATTATGATTTTT-----------CGATTTCTGA--T----------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTTTTTGTAAC---------GGGAAGA-CCAC----AACACCAACACACACA-CACACACACATACACATACTCACCTTACACTCAC-TTACAAAAACAG--------------------------------------CTGCA-------------------------------------------AACGTTA-----------------TAATGT---GC-C |
| droMoj3 | scaffold_6654:2211641-2211924 - | GATTCGTCACATTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGACGTAAGTCGAAAATGAACATTAACTACTGCACTCTTCTATGCAC---T---T------TT-------------------------------GCTAATGTCGCTTGT-----GT-------TTATC-------------------------------------------------------------------------------GATTTATGATTTATCGAGAAATATTCACAC-CCACCA---------------------------------------CACACACACACACA------TACCTACATACACACTCAAA--------------------------------------------CACATATATATT----TTA-AACTCGCTCTCACTCG------AACAGCTG------C---AAAAA--------------------------------------------------------------------------------AAAAAAATAAAT--AAATCAAACCAAAC--------TAC----------------------------------------AA--AAAAAAGTTA-----------------TAATGT---GC-C | |
| droGri2 | scaffold_15110:2695803-2696076 + | GATTTGTCACATTTGAGACGGAACAGGAGGCGCAAAGACTGCAGGCGGAGGTGAGTTCAA--------------------AGTTCATATGCTA------------------------------------------ATCTTGCTAATGACGCCTGT-----GTTA-ATTTCTATCGAT--TAATTATCG-------------------------------------------------------------------ATTATGATTTTA-----------CAATTTCTGA--T--GCTTTTTGTGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------CC----------------GGGAGGAGTCACACACACCACAAACACACACTCCACACATACACACACACACTCCACACACA--CACTCCACACATAC-----------------ACACA-------------------------------------------------------------CACACAACAAAAATAAGTATGAACTGTTATAATGT---GA-C | |
| droWil2 | scf2_1100000004837:1495835-1496107 + | GTTTCGTTACCTTCGAAACCGAACAGGAGGCTCAAAGACTGCAAGCAGATGTAAGTTC----------------------AATTAGTTACCAA---T---CCACGTT----------------------------AATTTGCTTATTATGATTAT-----CCCAAAT--TTCTTGGCATTG--T------------------AT-------AACAAAAAC--ACACATT-T----ATTTGAT---TTACCACAA-TTTTTGATTT-------------------TCGA--T---ATT-------------------------------------------------------------------------------TG-----------------------------------------------------------------------------------TCCGCAAG---------AAGAAGA-CGTC----GCCTT-----------------TGTC-----------------------------------------------------------------GTTCTCTTCGTCTTCTTGTCGTCGT---TTGCCAAACAAAAAAGAACTGTCA--TACTCTATCTGTTA---------------TCTTGT-----GC | |
| dp5 | XR_group8:8042784-8042903 - | TTGTTTTT---------------------------------------------------------------------------------GTTT---T---G------TT-------------------------------TTTAGCACAATTTGC------------------------------------------------------------------------------------------------------------------------------AT-TAGCAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TA-CACT----GAAAGAAAGAGACACA-CACACACACACACACGTACTCGATAAACAATATC-AAAATGGAAAGC--------------------------------------C-------------------------------------------------TTCAA-----------------AAG--T---GC-G | |
| droPer2 | scaffold_28:696086-696214 - | ATC--GATCCATTCAT----------------------------------------AA----------------------CGTTTTTTTGTTT---T---G------GT-------------------------------GCA--TTTAGTC-GTTAAATTG------------------------------------------------------------------------------------------------------------------CTTAAAC-ACA-CA---------------------------------------CACACACACACACA------CACATACACAAAGACTCAAA--------------------------------------------ATCACACA-------------------------------------------------------------------------------------------------------------------------CAT-CCACATGCACAT--------------------------------------------------------------------------------------------------------------CCACAT-----AC | |
| droAna3 | scaffold_13337:19509653-19509867 - | dan_2734 | GGTTCGTCACATTCGAGACGGAGCAGGAGGCACAAAGACTGCAAGCGGATGTAAGTAC----------------------CACTCCATTGGCC---TTCCG------TTCCGCTAATATATATCTCGCCTGTTGA----------------------AACGT-------TAA----------GAGTCGATTATGATTTCAAAAA-------AAAAAAAAC--AAAAAACA---------------------------------------------------------------------------------------------------------------------------------A---------GAA----TG----GAAGGACGGA--CTAT------------------------------------------TTT---TTG------TAGAG---------------------AAG-----------------------------------------------------------------------------AACACTTA--------------------------------------------------AAAAAA----------------------AAAA-----------------AATTGATACT-AC |
| droBip1 | scf7180000396360:704189-704430 - | GGTTCGTCACATTCGAAACGGAGCAGGAGGCACAAAGACTGCAGGCAGATGTAAGTAC----------------------CATTCCACTGACCACCTTCCG------TT--GCTAATA--TCTCTCGCCTGTTGA----------------------AATGT-------TTTTT--------------------------GC-----------------------------------------------------------------TGAAAAATATTTGATT-CCGAAAT--GTCGCCAGTAT----ATATTATTATTTAA----------------------------AAC-A----A---------AAATATTTGTGTAA-TCGACGGA--CTAT------------------------------------------------TT---------GTATTG------T---GTAGA-----------------------------------------------------------------------------------GAAACA----------------------------------------------------AAAACAAAAAAA-------------ATGTATA-----------------AATTGT---AC-T | |
| droKik1 | scf7180000302486:1611911-1612177 - | GGTTCGTCACATTCGAAACGGAGCAGGAGGCGCAAAGACTGCAAGCAGATGTAAGTAC----------------------CAGTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TAAAA--------------------------ACAA----------AAAAACCA--AAGAAAA----TTAAGAC--CGCGTCA--GTATTATGATTT-------------CGAAACCCGA--T--GAAGCCAGCG-------------------------------------------------------------------------------AACCAACGAA--CCTT---GAATCA-------------------TTTTAAAAACCC-TTCT-------------CGGG---------------------AG---------------------------------------------------------------------------A--------------------------GTACCCTACAGCAGCATATCGTATATGTAT------------------ATCCAACACACAACTATA-----------------AATTGT-----GC | |
| droFic1 | scf7180000454065:2564204-2564515 + | GATTCGTGACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTGAGTAC----------------------CTCTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AAACAAAACAAA--AAAAAGCAAAAAAAAAA----ATAAGAA---ATGCCA--GTATTATGATTT-------------CCATT-CCGA--T--GAAGCGAGCGTTTGAACATCACCA----------------------------ACAAAACCTACGAAC---------GAT----TG----AACGAACGAA--CCC--AACA-CATC---------------TT----TTA-GATTT-CTTTTGATTG------TCGCGTTA------T---GAAAA-----------------------------------------------------------------------------------GATACAA-----AACGA-----AAG--------TGCAGCA-------------T------------------ATGCAA--CACAACCTAA-----------------AATTGT-----GC | |
| droEle1 | scf7180000491193:3345915-3346239 - | GATTCGTCACATTCGAAACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAC----------------------CACTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AA-----------------------CAAAAA----TTAAGAA---ATGCCA--GTATTATGATTT-------------CGATT-CCAA--A--GAAGCGAGCGT--GAACATCATCAACCAACCAACCAACCAACCAACCAACCAAC--CACCTACAAAC---------GAT----TG----AACGAACGAA--CCTTATACA-CCTC---------------T--TATATA-GAGCA-TTTTCGATTG------TCGCGTCA------TAAAGAAAA-----------------------------------------------------------------------------------GATATAAAC--AAATCA-----CAC--------TGCAGCA-------------T------------------ATCCAA--CACAACTTAA-----------------AATTGT-----GC | |
| droRho1 | scf7180000777034:180806-181094 - | GATTCGTCACCTTCGAAACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTGAGTAC----------------------CACTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AAA--------------------------AA----CTAAGAA---ATGCCA--GTATTATGATTT-------------CGATT-CCGA--A--GAAGCGAGCGT--GAACATCACCA--------------------------CAAC--CACCTACGAAC---------GAT----TG----AACGAACGAA--CCT--CACA-CCTC---------------TT----ATA-TAGCT-TTTTCGATTG------TCGCGTTA------C---GAAAA-----------------------------------------------------------------------------------GATATAAAC--AAATCA-----CAC--------TGCAGCA-------------T------------------ATCCAA--TACAGCTTAA-----------------AATTGT-----GC | |
| droBia1 | scf7180000300910:2101037-2101307 + | GATTTGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGACGTGAGTAC----------------------CACTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTAAACGT-------TTGCC--------------------------ATAC----------------------AAGAAATGAATAAGAA--ACTGCCA--GTATTATGATTT-------------TGAAA-CCGA--A--GAAGCGATCGT--GAACATCACCA----------------------------AC--A--------AC---------GAC----TG----AAC---------CCC--ACCA-C-TC---------------GT----ATA-GAGCT-TTTTCAATTG------TCGCGTTA------C---GAAAA-----------------------------------------------------------------------------------GATATAAGC--CAACCA-----CAC--------TGC----------------------------------------AA--CACAACCTAA-----------------AATTGT-----GC | |
| droTak1 | scf7180000415356:81998-82299 + | GATTTGTCACCTTCGAAACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAA----------------------CACTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AAACAAAACAA------AAG--A----AAAA----ATGATAA--ACTGCCA--GTATTATGATTT-------------CGATG-CCGA--TGAGAAGCGATCTT--GAACATCACCA----------------------------AC--AACCTACGTAC---------GAT----TG----AACGAACGAA--CCT--AACAACCTC---------------TC----TTA-GAGCT-TTTCCAATTGTAACAATCGCGTTA------C---GAAAA-----------------------------------------------------------------------------------GATATATAC--AAATCA-----CAC--------TGC----------------------------------------AA--CACAACTTAA-----------------AATTGT-----GC | |
| droEug1 | scf7180000409467:309734-310011 - | GCTTTGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAC----------------------CACTCCTTTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTAAC--------------------------AAAA----------------------AAAAA----ATGAGAAAAACTGCCA--GTATTATGATTT-------------CGATT-CCGA--T--GAAGCGAGTGTGAAGAGATCACCA----------------------------ACAGAAACTACAAAC---------GAA----CG----AACGAACGAA--CTAT-ATGAACCT-------------------TGTTAA-GAGCT-TTTT-------------------T------C---GAGAA--------------------------------------------------------------------------------A--GATCTAAAC--AAATCA-----CGA---------AA----------------------------------------AA--CACTACCTAA-----------------AATTGT-----GC | |
| dm3 | chr3L:9105482-9105783 - | GATTCGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAG----------------------TACTCCATTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AAAAAAAGAAAA--AAAAAAAGAAACAACAA----ATAGGAA---ATGCCA--GTATTATAATTT-------------CAAT-----------GAAGC---CG---------AACAA----------------------------AC--AAACTACGAAC---------GAT----TT----AACGAACGAAAACCA--AACA-CTTCATATATACATATATATT----ATA-GAGCT-ATTTCGATTG------TCGCGTTT------C---GAAAA--------------------------------------------------------------------------------G--GATATGTACGAAAATCA-------------------------------------------------------------CACACAACTTAAAA---------------AATTGT-----GC | |
| droSim2 | 3l:8882202-8882504 - | dsi_1927 | GATTCGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAG----------------------TACTCCATTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACA--------------------------AAAA-------AAAAAAAAA--AAACAACAA----ATAGGAA---ATGCCA--GTATTATAATTT-------------TAAT-----------GAAGCGAGCGT--AAACATCACCA----------------------------AC--AACCTACGAAC---------GAT----TG----AACGAACGAAAACCA--AACA-CTTCATATA------TATATTTTATATA-GAGCT-CTTTCGATTG------TCGCGTTT------C---GAAAA--------------------------------------------------------------------------------G--GATATGTACGAAAATCA-------------------------------------------------------------CACACAACTTAAAA---------------AATTGT-----GC |
| droSec2 | scaffold_0:1367970-1368270 - | GATTCGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAG----------------------TACTCCATTGACC---T---G------TT-------------------------------GCTAATGTCGCCTGTTGAACGT-------TTACC--------------------------AAAA---GAAAA--AAAAAA--CAACAACAA----ATAGGAA---ATGCCA--GTATTATAATTT-------------TAAT-----------AAAGCGAGCGT--AAACATCACCA----------------------------AC--AACCTACGAAC---------GAT----TG----AACGAACGAAAACCA--AACA-CTTCATATA------TATATT----ATA-GAGCT-CTTTCGATTG------TCGCGTTT------C---GAAAA--------------------------------------------------------------------------------G--GATATGTACGAAAATCA-------------------------------------------------------------CACACAACTTAAAA---------------AATTGT-----GC | |
| droYak3 | 3L:2812194-2812283 + | GATTCGTTACCTTTGAGACGGAGCAGGAGGCGCAAAGACTGCAAGCGGATGTAAGTAG----------------------TACTCCAATGAAC---T---G------TT-------------------------------GCTAATGTCGCCTGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4784:4938955-4939042 + | GATTCGTCACCTTCGAGACGGAGCAGGAGGCGCAAAGACTGCAGGCGGATGTAAGTAG----------------------TACTCCAATGACC--------------TT-------------------------------GCTAATGTCGCCTGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:07 AM