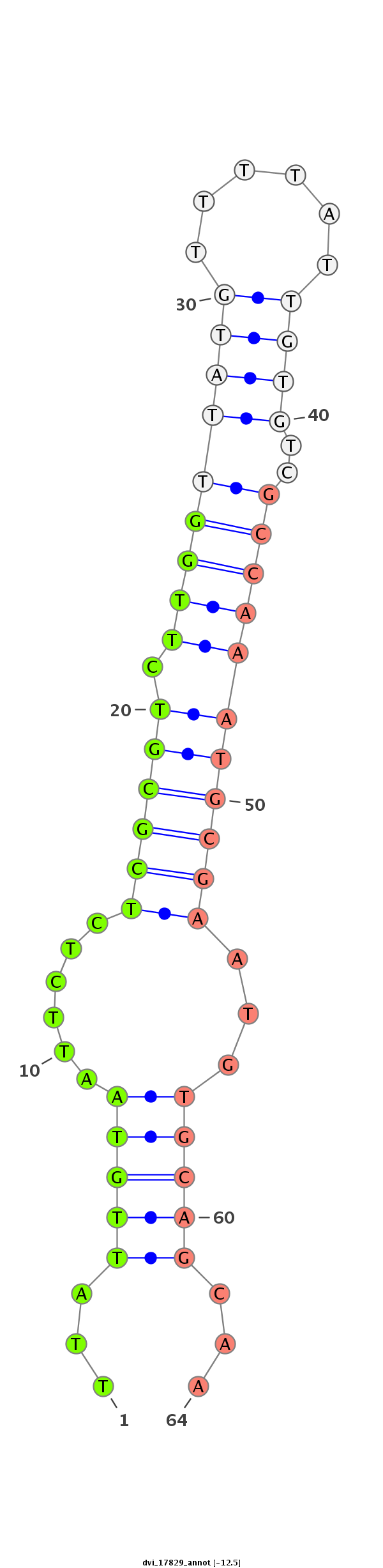
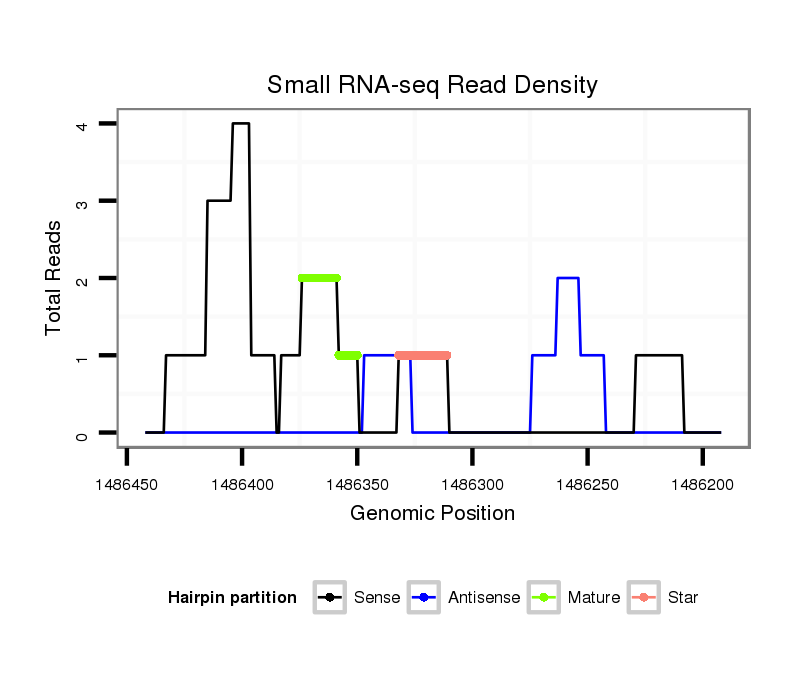
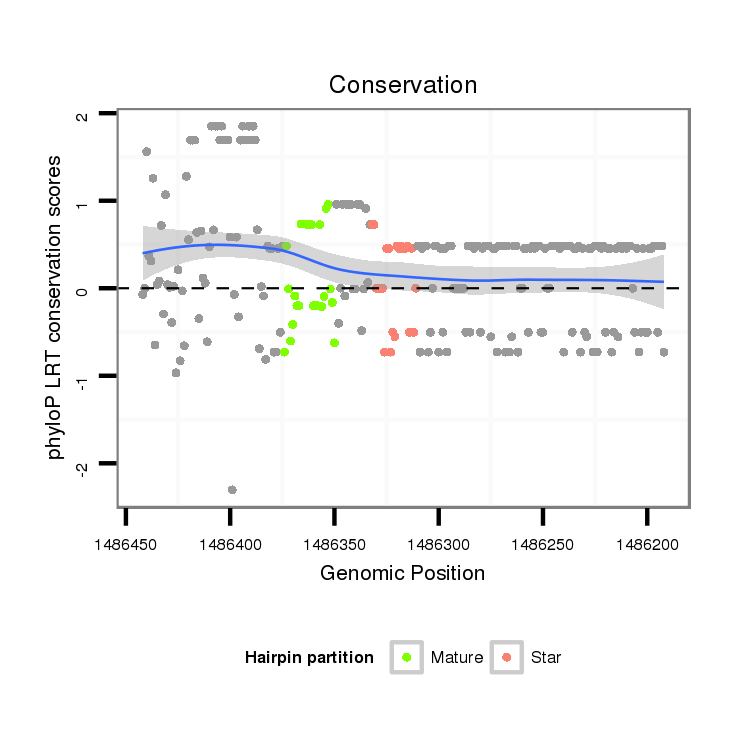
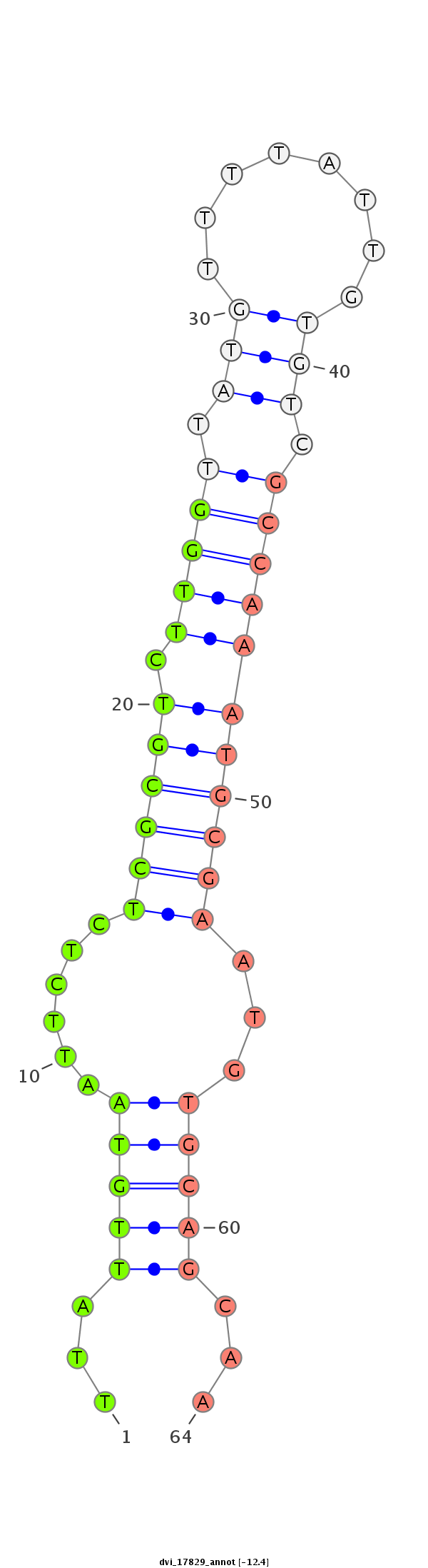
ID:dvi_17829 |
Coordinate:scaffold_13049:1486242-1486392 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.4 | -12.0 | -12.0 |
 |
 |
 |
exon [dvir_GLEANR_12569:1]; CDS [Dvir\GJ12612-cds]; intron [Dvir\GJ12612-in]
No Repeatable elements found
| -----------------------------------###############--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GAGAGCCGCAGAGCGAAACGAGGCGCTCAACTGAAATGAGCGCGGCGGAGGTGAGTGCGATGCGACATTTATTGTAATTCTCTCGCGTCTTGGTTATGTTTTATTGTGTCGCCAAATGCGAATGTGCAGCAATTCAATTAAATGCGAATTAAATCGAATCTTCGTAATCCCAAAGAATATGCGCGTGTGTATGGTATTATGTTATTACGTGCATCCATATGTGCCGACAAGTGTTACGACGCGCCAAGAAT ********************************************************************...(((((......((((((.(((((((((......))))..)))))))))))...)))))...*********************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M028 head |
M047 female body |
SRR060654 160x9_ovaries_total |
SRR060657 140_testes_total |
SRR060667 160_females_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060683 160_testes_total |
SRR1106720 embryo_8-10h |
SRR060658 140_ovaries_total |
SRR060673 9_ovaries_total |
SRR060675 140x9_ovaries_total |
SRR1106726 embryo_14-16h |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................CAACTGAAATGAGCGCGGC............................................................................................................................................................................................................. | 19 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TGCGAATTAAATCGAAG............................................................................................ | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TTATTGTAATTCTCTCGCGTCTTGG.............................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................TCCATATGTGCCGACAAGTGT................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCCAAATGCGAATGTGCAGCAA....................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ATGCGACATTTATTGTAATTCTCTC....................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGCGCGGCGGAGGTGAGTG.................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........AGAGCGAAACGAGGCGCT................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................CGAGCGCGTCGGAGGTGA..................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CGCGTGTGTATGATATTGTG.................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GCCAACTAGTATTACGACGC......... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................TAGTATTACGACGCGCC...... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................AAGGAGCAAGGCGGAGGTG...................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CTCTCGGCGTCTCGCTTTGCTCCGCGAGTTGACTTTACTCGCGCCGCCTCCACTCACGCTACGCTGTAAATAACATTAAGAGAGCGCAGAACCAATACAAAATAACACAGCGGTTTACGCTTACACGTCGTTAAGTTAATTTACGCTTAATTTAGCTTAGAAGCATTAGGGTTTCTTATACGCGCACACATACCATAATACAATAATGCACGTAGGTATACACGGCTGTTCACAATGCTGCGCGGTTCTTA
***********************************************************************************************************************...(((((......((((((.(((((((((......))))..)))))))))))...)))))...******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060656 9x160_ovaries_total |
SRR060673 9_ovaries_total |
SRR060688 160_ovaries_total |
SRR060689 160x9_testes_total |
GSM1528803 follicle cells |
V053 head |
SRR1106728 larvae |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................CTGTTCACGATGCTGCGCGGT..... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................ACGCGCACACATACCATAATA................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TACAAAATAACACAGCGGTTT....................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GGGTTTCTTATACGCGCACAC.............................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGCATGCCATAATACAAAAAT............................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................AATGCTGCGAGGTTCTTA | 18 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TCAAAATGATGCCCGGTTCTT. | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................AATGCTGCGAGGTTCT.. | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................GCTGCGAGGTTCTTA | 15 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........CTCGCGCTTTGCTCCGAGA................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................ACGAGAATGCACGTAGGTG................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13049:1486192-1486442 - | dvi_17829 | GAGAGCCGCA----GAGCGAAACGA-G-GCGCTCAACTGAAATGAGCGCGGCGGAGGTGAGTGCGAT-GCGACATTTATTGTAATTCTCTCGCGTCT----TGGTTATGTTTTATTGTGTCGCCAAATGCGAATGTGCAGCAATTCAATTAAATGCGAATTAAATCGAATCT--TCGTAATCCCAAAGAATA----------TGCGCGTGTGTATGGTATTATGTTATTACGTGCA-----TCCATATGTGC--C--------GACAAGTGTTACGACGCGCCAAGAAT |
| droMoj3 | scaffold_6654:2336157-2336380 + | GAGCTCG-CA----GAGCACTAAAC--TTCGCTCAGCTGAAATGAGCGTCGCTGAGGTGAGTCCGCTTGCTCCGTGTAGAG--------------------------------------C-------GGCTGTTGTGCAACG-TGCAAGC-AAGGTGCAT------AAGTCTTCTTGTAAACCAAAATACAAATCGAAATTTTT--CGCGTGTACAG--TTATGTGATTGCGTTCGCTGTGACCCTCTGTACGTCGTAGCATTTGCTAGTGTT-TGCTGTACCAAAAAG | |
| droGri2 | scaffold_15110:2589640-2589723 - | GCGAGCAGCAATAATAAAACGAAATTG-GCGCTCAG-TGAGATGAGCGCAGCGGAGGTGAGAC------------------------------GTCT----TATTAATGTTTTATTTTGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | XR_group8:2184841-2184896 - | AAGAGCGAG------AGCGCTGCGT-GTGCGCATTGTCGAAATGAGCGCGACAGAGGTGAGTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_24:872597-872652 + | AAGAGCGAG------AGCGCTGCGT-GTGCGCATTGTCGAAATGAGCGCGACAGAGGTGAGTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396641:1191195-1191233 - | GAG------------------------TGCGCAGAGCGAAAATGAGCGCTGTCGAGGTGAGTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453839:1884205-1884302 - | GAGAGCGAC-------------TGA-GTGCGCAGAGTAAAAATGAGCGCCGCAGAGGTGAGTATTT-----------TGTTCGATTCTCCTACCGCTAAATAGTTTGTTTTTATTTGAGTCGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779409:176001-176042 + | ACGAG----------------------TGCGCATAGTGAAAATGAGCGCTACAGAGGTGAGTGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4784:17325415-17325455 + | A--------------------GCGA-GTGCGCATAATAAAAATGAGCGCGACAGAGGTGAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 03:26 AM