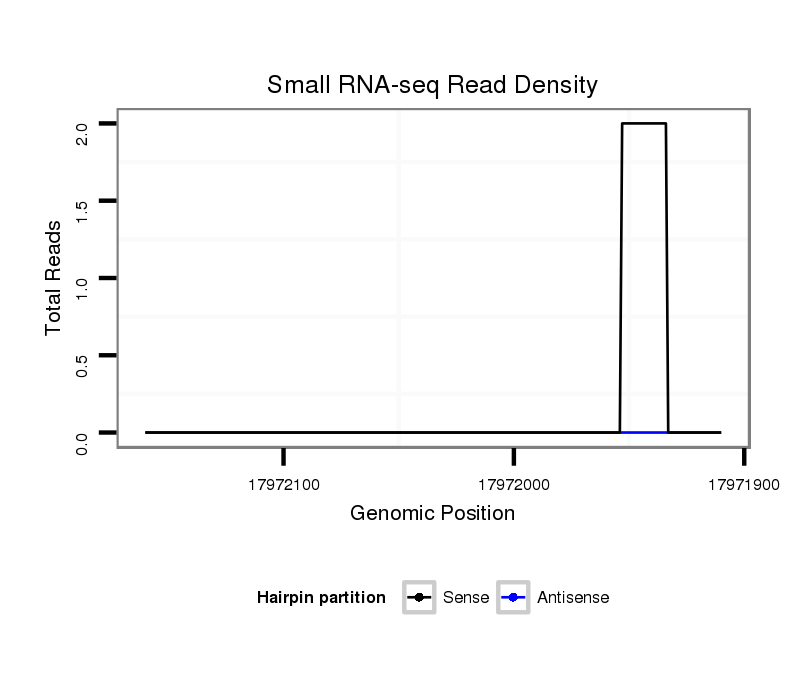
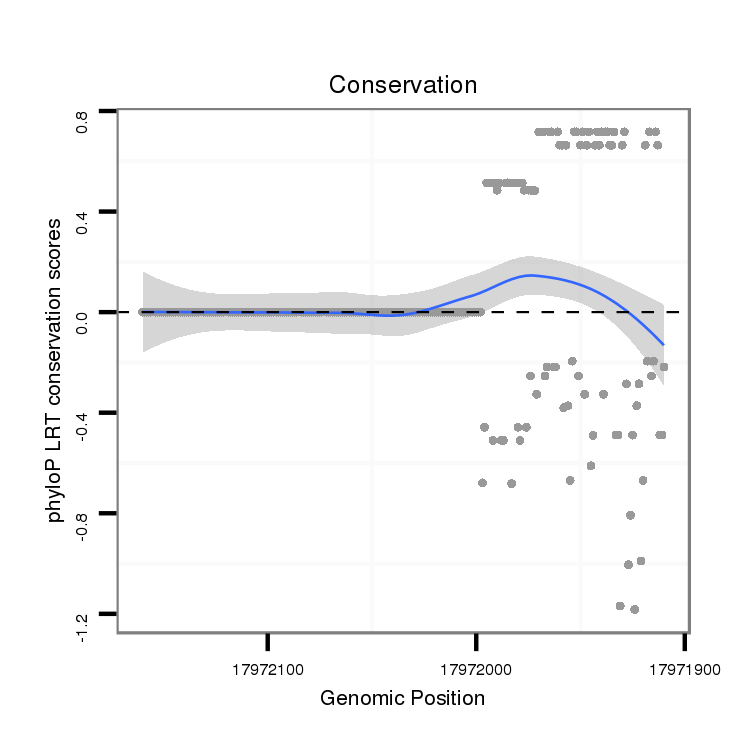
ID:dvi_17427 |
Coordinate:scaffold_13047:17971960-17972110 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_8005:2]; CDS [Dvir\GJ22622-cds]; intron [Dvir\GJ22622-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TACATATGTATCTACCAATAGCATATATTCAGAGCTTTTTATATATATTTACTTTTTGACTTATTAATATGATATTTTTGGTAACACTTGTAATTGCAAGTTGATCTCGATATCCAATAAATAAAATATCAGCTGCTATTCGAAATATATTCAGATAAACTATACAAATTGTTAAATTAAGTTGCCTTACAATTCTTGCAGGAGGCCAAAGTGGAGACAGAGATGGAGTTCATACTTGTACGCAACTCGCC **************************************************((((((((((..(((..((((.....(((((......(((.(((.((.(((((((......................))))))))).)))))).......))))).....))))..))).))))))..))))...................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060657 140_testes_total |
V116 male body |
SRR060673 9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................AAAGTGGAGACAGAGATGGA........................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AAAGTATCAGCAGCTATTCG............................................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................AAATATCAGCTGGTATTCT............................................................................................................. | 19 | 2 | 10 | 0.20 | 2 | 0 | 0 | 1 | 1 | 0 |
| ...........................................................................................................................................................................................................................GAGATGGAGATGATACCTGT............ | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................ATACATTCAGATAAGCTA......................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
ATGTATACATAGATGGTTATCGTATATAAGTCTCGAAAAATATATATAAATGAAAAACTGAATAATTATACTATAAAAACCATTGTGAACATTAACGTTCAACTAGAGCTATAGGTTATTTATTTTATAGTCGACGATAAGCTTTATATAAGTCTATTTGATATGTTTAACAATTTAATTCAACGGAATGTTAAGAACGTCCTCCGGTTTCACCTCTGTCTCTACCTCAAGTATGAACATGCGTTGAGCGG
**************************************************((((((((((..(((..((((.....(((((......(((.(((.((.(((((((......................))))))))).)))))).......))))).....))))..))).))))))..))))...................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060679 140x9_testes_total |
SRR1106726 embryo_14-16h |
M047 female body |
SRR060654 160x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................CCTCTGTCTCTACCTAAACTAT................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTCAACTGGAGCTTTAGGTTGT.................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................TTTCTCGTCGACGATAACCTTT.......................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................ACCTCAAGTAGGAACATGTT........ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................AAAAAAACTATTGTGATCATT.............................................................................................................................................................. | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:17971910-17972160 - | dvi_17427 | TACATATGTATCTACCAATAGCATATATTCAGAGCTTTTTATATATATTTACTTTTTGACTTATTAATATGATATTTTTGGTAACACTTGTAATTGCAAGTTGATCTCGATATCCAATAAATAAAATATCAGCTGCTATTCGAAATATATTCAGATAAACTATACAAATTGTTAAATTAAGTTGCCTTACAATTCTTGCAGGAGGCCAAAGTGGAGACAGAGATGG--AGTTCATACTTG-TACGCAACTCGCC |
| droMoj3 | scaffold_6540:13738231-13738296 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------C----------------------CTAAAATCTTTATAGGAGAACAAGGTTGACCCAGATATGG--AGGCCACCGGGA-CTAGCAGCTCGCT | |
| droGri2 | scaffold_15074:6670151-6670239 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAAAATGTATAATGAAAATGTCT--CAATTCTTGCAGGCGAATAAAGTGGAGCCAGAGATGGCTACGGCACTGGCAGCCAGTAATTCCGC | |
| droEug1 | scf7180000409802:6379-6379 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------C------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/16/2015 at 11:29 PM