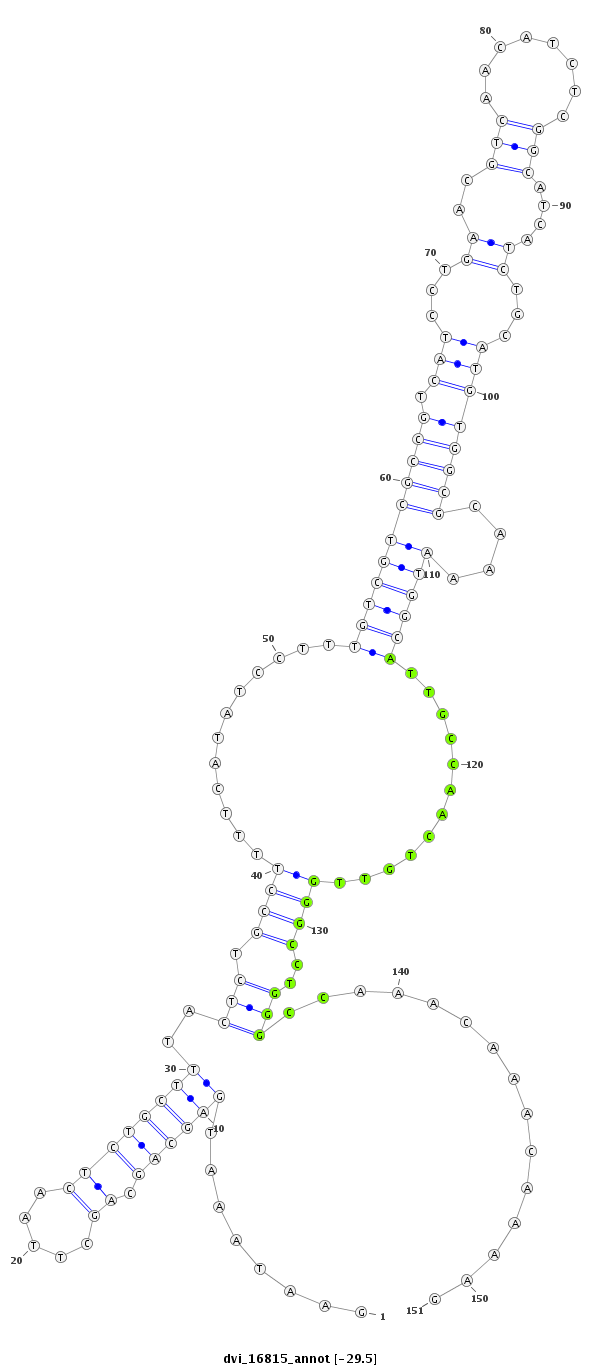

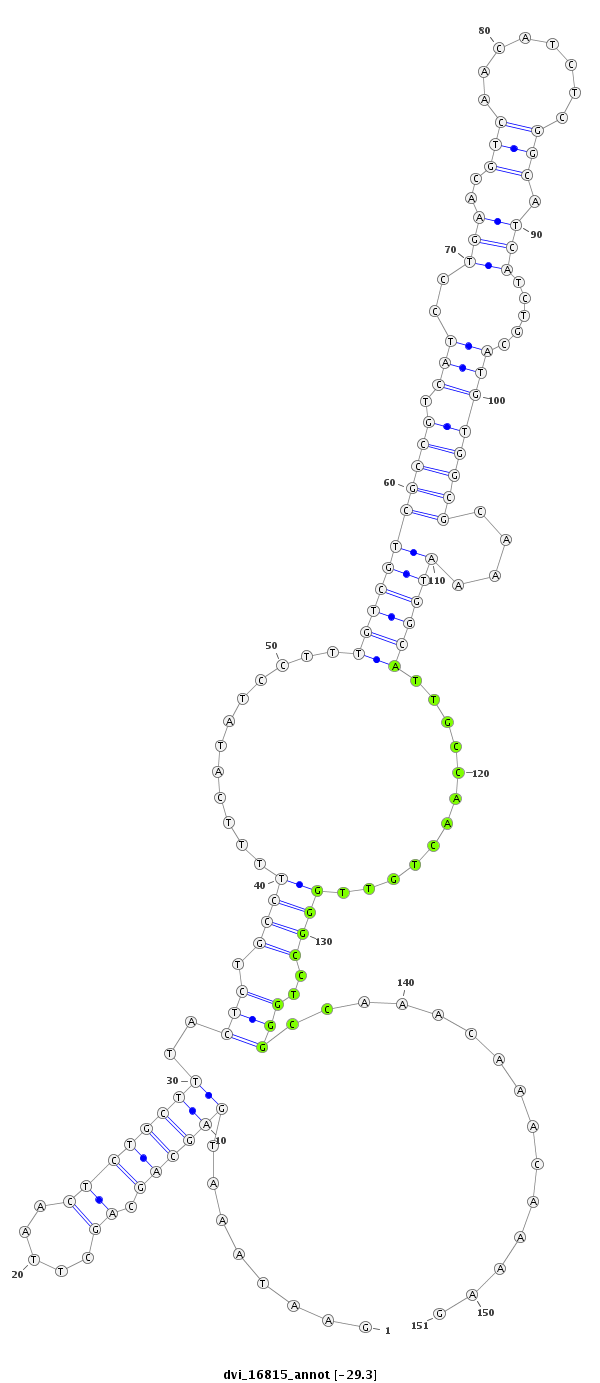
ID:dvi_16815 |
Coordinate:scaffold_13047:13514928-13515078 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.3 | -29.2 | -29.0 |
 |
 |
 |
exon [dvir_GLEANR_8231:10]; CDS [Dvir\GJ22866-cds]; intron [Dvir\GJ22866-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGTATCTAAAGAAGAAAAGTGCGAGAAGGGCCTGATGCAGACAATAGCGAGAATAAATGAGCAGCAGCTTAACTCTGCTTTACTCTGCCTTTTCATATCCTTTGTCGTCGCCGTCATCCTGAACGTCAACATCTCGGCATCATCTGCATGTGGCGCAAAATGGCATTGCCAACTGTTGGGCCTGGGCCAAACAAACAAAAGACCTACATGACATACGCACCGACGCCCAACTGCAGCTACGGTGGCTTACC **************************************************........((((((.((.....))))))))..(((.((((............(((((((((((.(((...((..(((........)))....))...))))))))....))))))............))))..)))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060655 9x160_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060657 140_testes_total |
M047 female body |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....CTAAAGAAGAAAAGTGCGAGAAGGGCCT.......................................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................ATTGCCAACTGTTGGGCCTGGGCC............................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CATCCCGTGCGTCAACATCT..................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................TCGGCACCGTCACCCTGAA................................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................GTGCGAGAAGGGCGTGAGGG..................................................................................................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................AATCTCGGCATCACCTCCA....................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCATAGATTTCTTCTTTTCACGCTCTTCCCGGACTACGTCTGTTATCGCTCTTATTTACTCGTCGTCGAATTGAGACGAAATGAGACGGAAAAGTATAGGAAACAGCAGCGGCAGTAGGACTTGCAGTTGTAGAGCCGTAGTAGACGTACACCGCGTTTTACCGTAACGGTTGACAACCCGGACCCGGTTTGTTTGTTTTCTGGATGTACTGTATGCGTGGCTGCGGGTTGACGTCGATGCCACCGAATGG
**************************************************........((((((.((.....))))))))..(((.((((............(((((((((((.(((...((..(((........)))....))...))))))))....))))))............))))..)))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
GSM1528803 follicle cells |
V053 head |
M047 female body |
SRR060661 160x9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
M027 male body |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................TGATTGGCTGCAGGTTGACG................. | 20 | 3 | 3 | 1.33 | 4 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTTGGGTGCGGGTTGACG................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TAGAGCCGTAGTAGACGTACAC................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CGTGGAATTGAGATGCAA.......................................................................................................................................................................... | 18 | 3 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CGGGTGGGCGTCGAAGCCACC...... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................GAATGGGTGGGTGCGGGTTGA................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CAGTTGTAAGGCCGTAGT............................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GCTCTTGACGGACTACGACT.................................................................................................................................................................................................................. | 20 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TGGCTGCAGGTTGACGACAA............. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CGTCGAATTGAGATGCAAG......................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGACGTCGTTGCAACTGAAT.. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GCCGTGGAATTGAGACGCA........................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGAGTGGGTGCGGGTTGA................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................AGTAGAAATTGCAGTTCTAG...................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TTTCGTCTTTTCACTCCCTTC............................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TGCAGTAGTACTTGCAATTG......................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGCAGGTTGACGACAATGC.......... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................GGGTGAACGTCGATGCCAA....... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................CGTGGAATTGAGATGCAAT......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGATTGGCTGCAGGTTGAC.................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
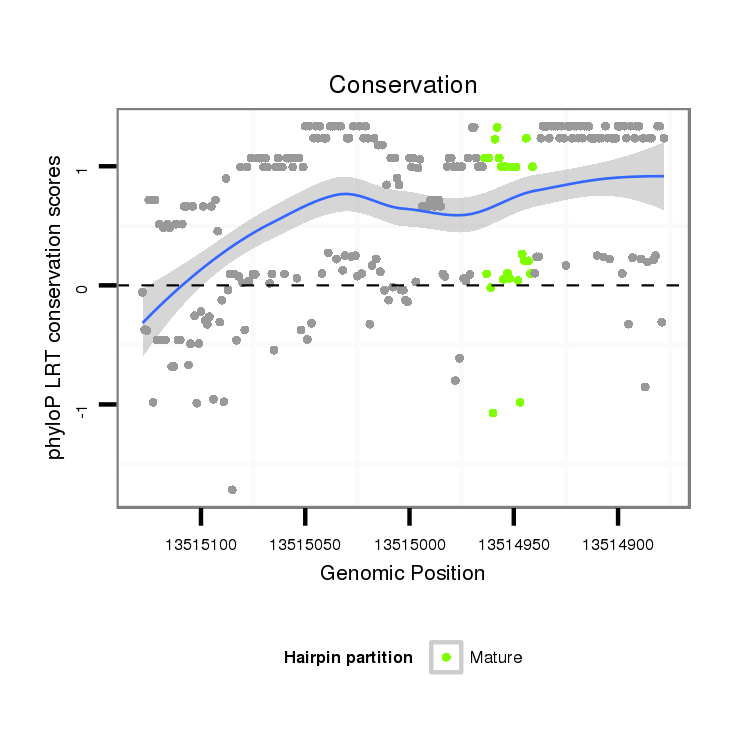
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:13514878-13515128 - | dvi_16815 | CGTATCTAAAGAAGAAAAG---TGCGA-------------------------------------------------------------GAAGGGCCTGATGCAGA----------------------------------------CAATAGCGAGAATAAATGAGCAGCAGCTTAACTCTG--C-----TTTACTCTGCCTTTTCATATCCTTTGTCGTCG--CCGTCATCCTGAACGTCAACAT--CTCGGCAT--CATCTGCATGTGGCGCAAAATGGC-ATTGCCAACTG-------------------TTGGGCCTGGGCCAAACAAACAAAAGACC---TACATGACATACGCACCGACGCCCAACTGCAGCTACGGTGGCTTACC |
| droMoj3 | scaffold_6540:10754767-10755103 - | CATATTT----------------GCTCGTATGCTACGTACATTTTTTCTGATGGCCACCGATTGGGGGGCGGTGAGCAAGGTGAGCAAGGTGAGCAAGGTGAACACGGTGAGCCAGGTATCTGGTGTCTGGGCCTGCGTCATTTTCAATTGCAGGAATAAATGAGCAGCAGCTTAACTTTA--CTTTACTGTGCTCTGCCCTTTCATATCCTTTGTCGTCA--CCG------TCA------ACAA--CTCGGCATCTCATCTGCATGCGGCACAAAATGGC-ATTCGCAACCG-------------------TCGGGCATGGGCCAAACAAACAAAAGACCTCCTACATGACATACGCACCAACGCCCAACTGTAGCTACGCTGGCTTAAC | |
| droGri2 | scaffold_15074:507550-507811 + | CAGATATGAGGGAGCCAGATCTTGCTC-------------------------------------------------------------GACCGGACTGTT-------------------------------------GTCATTTCGAATTGCAAGAATAAATGAGCACCAGCTTAACTCTG--CTTTACTTTGCTCTGCCTTTTCATATCCTTTGTCGTCA--CCGTCATCCTGAACGTCAACAA--CTCGGCAT--CATCTGCATGCGCCGTAAAATGGC-ATTGCCAACTG-------------------TTGGGTTATGTCCAAACAAACAAAAGACC---TACATGACATACGCACCGACGCCCAACTGTAACTACGCTGGCTTATC | |
| droWil2 | scf2_1100000004943:13731212-13731370 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTTCTCTGCCTTTTTATGCCCTTCATCGTCA--ACATTA--------------------------------CACATGGGCCGCATAATGGCGACTGGCAACTGGCAACTGGCAACTTGCACGA------TGGGGCTAACAAACAAAAGACA---TACATGACATACGCGCCGACACCCAACTGTAGCTATGGGGGCATACC | |
| dp5 | 2:25079016-25079203 + | G----------------------------------------------------------------------------------------------------CCAA----------------------------------------CGCCAACAGGCACTAATGAGAGCCAGCCTAACTCTATGCCCCACTCTGCTCTCCCTTTTCATGTCCATTATGGTCGCGCCGTCACCATGAACACACACAA------------------------------AA----------CA-------------------CACGATGGGCATGGGC-TGGCAAACAAAAGACC---TACATGACATACGCACCGACGCCCAAGTGCAGCTACGCTGGTTTACC | |
| droPer2 | scaffold_6:379015-379202 + | G----------------------------------------------------------------------------------------------------CCAA----------------------------------------CGGCAACAGGCACTAATGAGAGCCAGCCTAACTCTATGCCCCACTCTGCTCTCCCTTTTCATGTCCATTATGGTCGCGCCGTCACCATGAACACACACAAAACT-----------------------------------------------------------CACGATGGGCATGGGC-TGGCAAACAAAAGACC---TACATGACATACGCACCGACGCCCAAGTGCAGCTACGCTGGTTTACC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/16/2015 at 11:33 PM