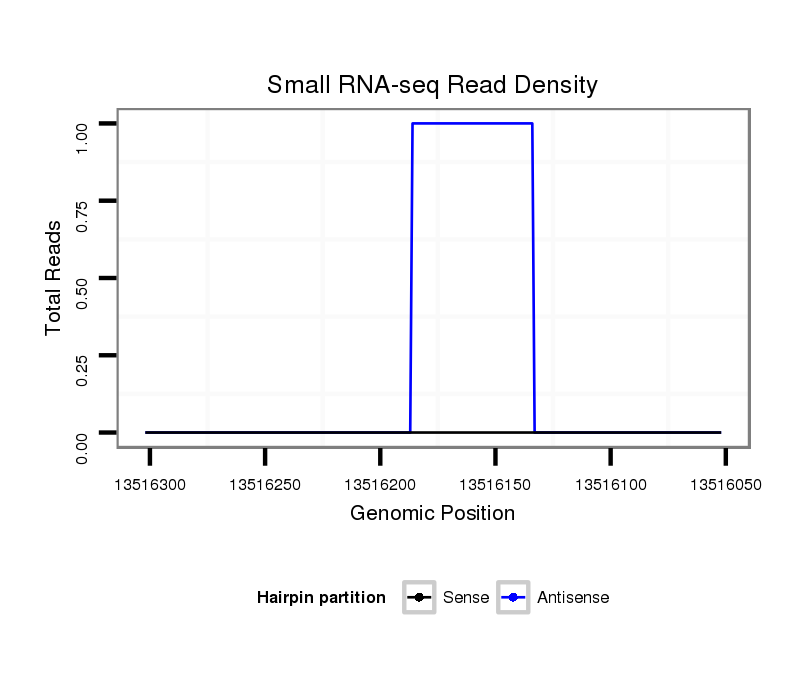
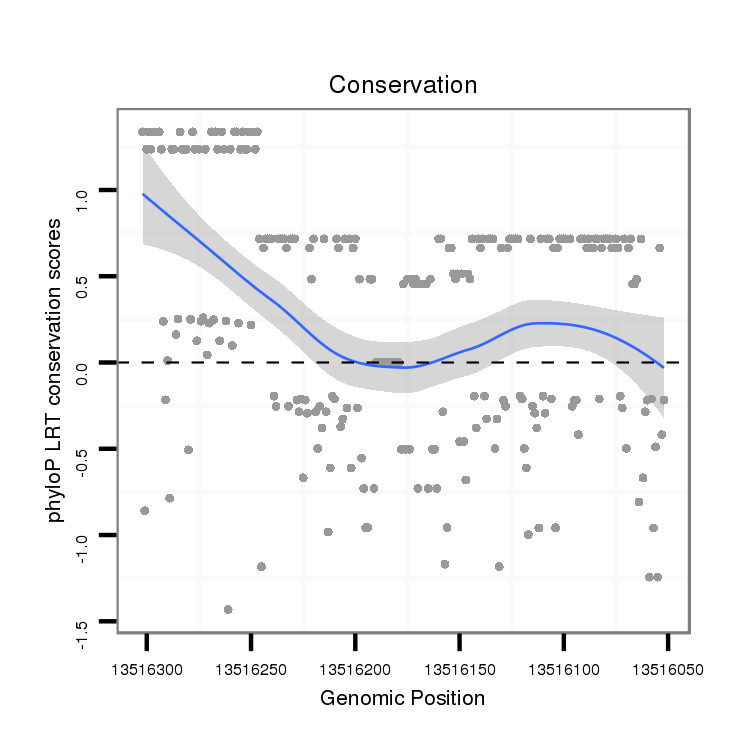
ID:dvi_16807 |
Coordinate:scaffold_13047:13516102-13516252 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_8231:9]; CDS [Dvir\GJ22866-cds]; intron [Dvir\GJ22866-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTGACTTAACAACGCCGGTGCCTTTGTCCAGTTTATCATCACGGAATGAGGTGAGTTTCAATTGAAATTGTTTAATACGGCATATTATAGGCATGGGTATGGATATGGGTTAAATAAGAAAGAGCGTAAACTGGGGCCTAGCATAAAGCTCAATCCTGTCTACTGGTTATCAGTACAATTACATGGAACTTAGCATACAGTAATTAACTTAATCACCTTTCAATAGCTGCAATGAGCTGTCTACATAGCAA **************************************************.(((((((((..((.(((((..........((((((.........))))))((((.(((((..........((((.....((((...))))......)))))))))))))(((((.....)))))))))).))))))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
V116 male body |
GSM1528803 follicle cells |
SRR060666 160_males_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
V053 head |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060667 160_females_carcasses_total |
M047 female body |
SRR060671 9x160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
M061 embryo |
SRR060663 160_0-2h_embryos_total |
V047 embryo |
SRR060685 9xArg_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
SRR060689 160x9_testes_total |
SRR1106727 larvae |
SRR060658 140_ovaries_total |
SRR1106714 embryo_2-4h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................CAGCTTATCACCCCGGAAT............................................................................................................................................................................................................ | 19 | 3 | 8 | 18.13 | 145 | 20 | 16 | 0 | 11 | 14 | 13 | 4 | 10 | 9 | 1 | 7 | 3 | 6 | 6 | 6 | 2 | 3 | 5 | 3 | 0 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| .............................AGCTTATCACCCCGGAAT............................................................................................................................................................................................................ | 18 | 3 | 20 | 2.40 | 48 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 4 | 0 | 0 | 1 | 10 | 7 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| ............................................................................................................................................................................................................................CAATAGCCGCAATGTGCCGTC.......... | 21 | 3 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CAGCTTATCATCCCGGAAT............................................................................................................................................................................................................ | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CAGTTTATCACCCCGGAAT............................................................................................................................................................................................................ | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................CAGAGCGTACACTGGGGCCT................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CAGCTTATCACCACGGAAT............................................................................................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TTAACAACGCCGGTGTAT.................................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ACTCAATGCTGTCTACTG...................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TAACAACGCCGGTGTATCT.................................................................................................................................................................................................................................. | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CAGCTTATCACCACGGAATT........................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CCAATAGCTGTAATGAGCTA............ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TAATTAACTCAATCCCCTGT............................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GTTAACAACGCTGGTGTCT.................................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TAAGCATGGTTATGGATAT................................................................................................................................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................CATGAGCTGTCTTCACAGCA. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATTAACAACGCCGGTGTAT.................................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CGTAGGCTGGGGCCTCGCA............................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CGGGATATTGTGGGCATGG........................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AACTGAATTGTTGCGGCCACGGAAACAGGTCAAATAGTAGTGCCTTACTCCACTCAAAGTTAACTTTAACAAATTATGCCGTATAATATCCGTACCCATACCTATACCCAATTTATTCTTTCTCGCATTTGACCCCGGATCGTATTTCGAGTTAGGACAGATGACCAATAGTCATGTTAATGTACCTTGAATCGTATGTCATTAATTGAATTAGTGGAAAGTTATCGACGTTACTCGACAGATGTATCGTT
**************************************************.(((((((((..((.(((((..........((((((.........))))))((((.(((((..........((((.....((((...))))......)))))))))))))(((((.....)))))))))).))))))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060662 9x160_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................GGAAGCAGGCCAAATAGTGG................................................................................................................................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TCTTTCTCGCATTTGACCCCGGATCGT............................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................ATTTCGAGTTAGGACAGATGACCAAT.................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................CCAATAGTCATGCTGACGTA................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......ACTGGTGCGGCCACGGAA................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................GAAGTTATAGACCTTACTCG.............. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:13516052-13516302 - | dvi_16807 | TTGACTTAACAACGCCGGTGCCTTTGTCCAGTTTATCATCACGGAATGAGGTGAGTTTCAATTGAAATTGTTTAATACGGCATATTATAGGCATGGGTAT-------GGATATGGGTTAAATAAGAAAGAGCGTAAACTGGGGCCTAGCATAAAG-------------------------------CTCAATCCTGTCTACTGGT--------TATCAGTACAATTACA-TGGAAC-TTAGCATACAGTAATTAACTTAATCACCTTTCAATAGCTGCAATGAGCTGTCTACATAGCAA |
| droMoj3 | scaffold_6540:10756059-10756316 - | TTGACTTAACAGCTCCGATGCCATTGTCTAGTCTATCATCACGGAACGAGGTGAGTTGCAATTGGAATTGCTTAACAAAGCATGGCATGACCATGATTTT-------CGAAAATCCTTC------------TGCAGACTTGGGCATGAAATGGGGTTCAACTCGGGTTTAGCTTAGTC---GGGTGC---------TCTACTGTT--------TAGACGTGCAATTACA-GCTAGC-TAAGCATACGGTAATTAGTTAAATCACCTTTCAATAGCTGCTAGGAGCTCTACAAAACTCTA | |
| droGri2 | scaffold_15074:506420-506676 + | TGGACTTAACCGCCCCCATGCCATTGTCCGGCCTATCATCAAGGAACGAGGTGAGTTCCAATTAAAATTGTTTATCGAGTC-TGTTCTGTGTGTGAGTATGCAGTTTGGAT--------------------------------------ATGCTGTTCTATTCACGGTTAGTTTTGAATGTTAGAGCTCAGTTATGTTGACTAGTTATGTAGTTATCGGAACAATTATGATGAAAAGGCAAAATGCTGTAATTAACAAAATCACCTTCCAATAGCTGTAATGA---CTACTGGCCCCTT | |
| droWil2 | scf2_1100000004943:13729737-13729792 + | TAGACTTAACAAGTCCGATGCCCCTGCCCAGCCTTTCGTCACGCAACGAGGTGAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | 2:25077322-25077377 + | TCGACTTAACAGGTCCGATGCCATTGCCCAGCCTATCGTCTAGGAACGAGGTAAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_6:377429-377484 + | TCGACTTAACAGGTCCGATGCCATTGCCCAGCCTATCGTCTCGGAACGAGGTAAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/16/2015 at 11:35 PM