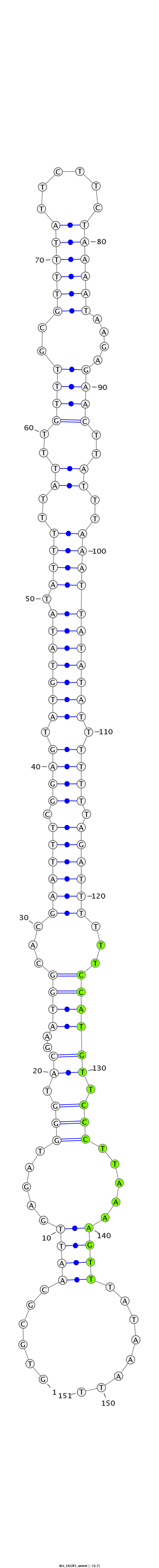
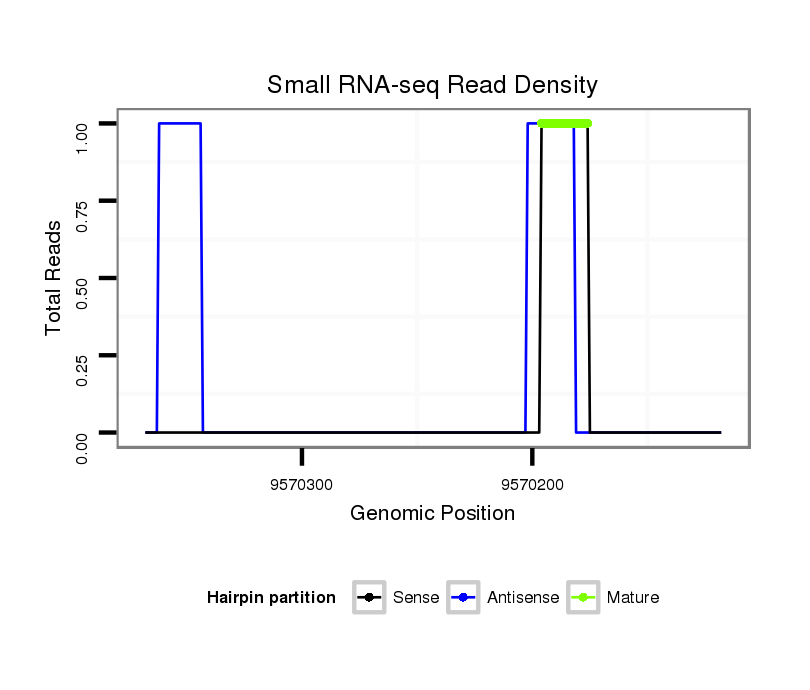
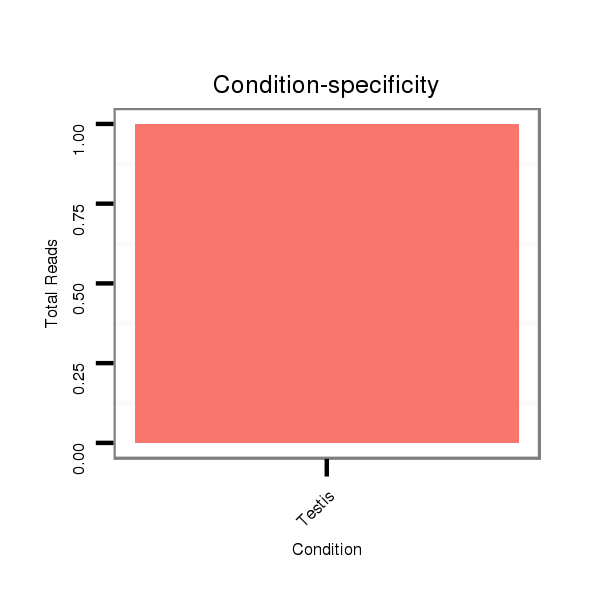
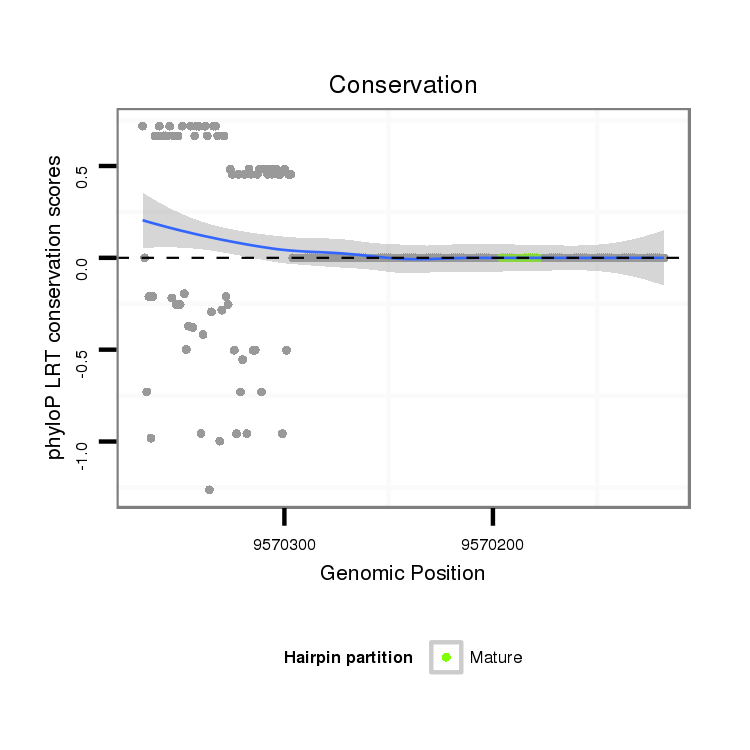
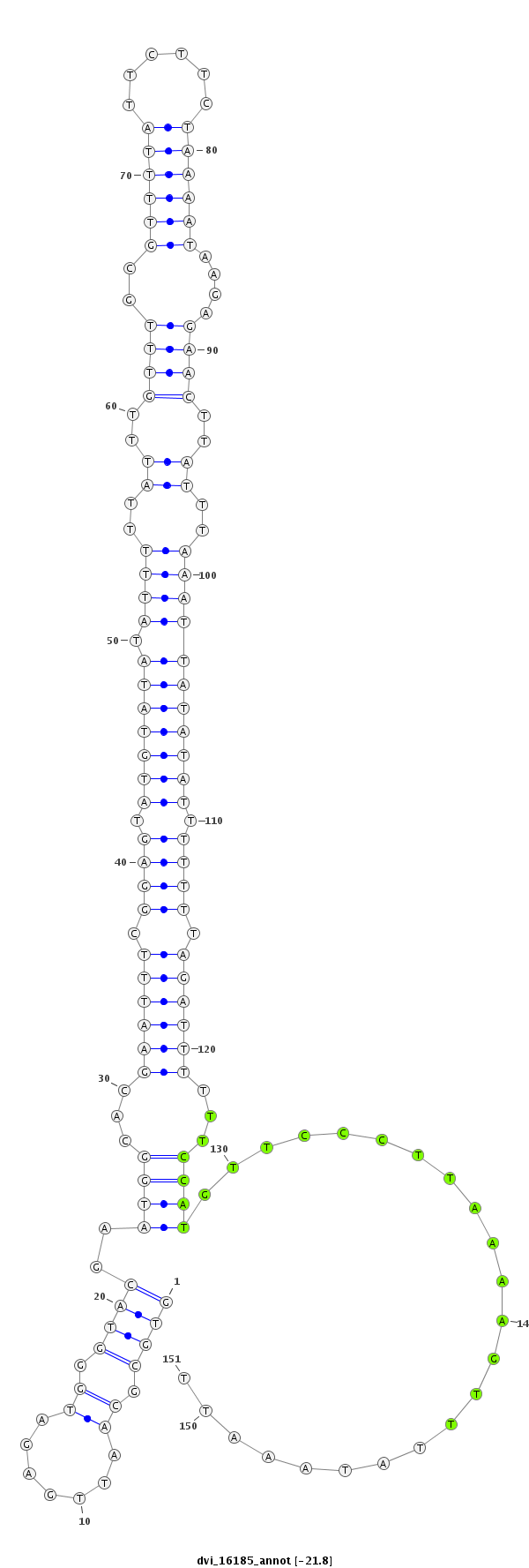
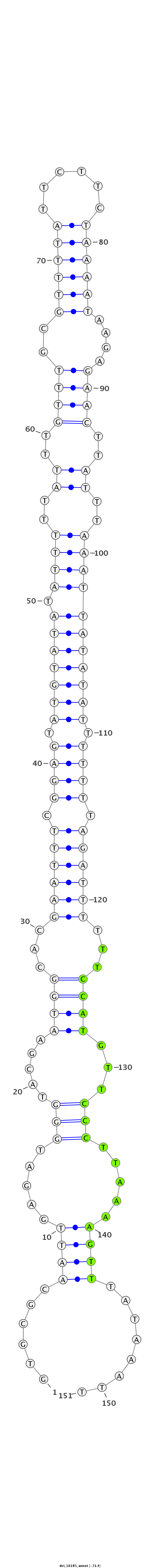
ID:dvi_16185 |
Coordinate:scaffold_13047:9570168-9570318 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.8 | -21.5 | -21.4 |
 |
 |
 |
exon [dvir_GLEANR_8453:1]; CDS [Dvir\GJ23092-cds]; intron [Dvir\GJ23092-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ----------------------------------################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTTAGACCAGGGCTGCACTAGACAACAAAAAGCGATGGACATTGCGCAAGGTGCGCAATTGAGATGGGTACGAATGGCACGAATTTCGGAGTATGTATATATTTTTATTTGTTTGCGTTTTATTCTTCTAAAATAAGAGAACTTATTTAAATTATATATTTTTTTAGATTTTTTCCATGTTCCCTTAAAAGTTTATAAATTATGTTATAAAATTCTTTGATATCGTTCTAAATCGTGGAATCAATATTTTA **************************************************......((((.....(((.((..((((...((((((.((((.(((((((.((((..((..((((..((((((......))))))....))))..))..))))))))))).)))).))))))...)))))).))).....))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060667 160_females_carcasses_total |
SRR060678 9x140_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060671 9x160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
M047 female body |
M061 embryo |
SRR060661 160x9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................CAATATAGATGGATACGAAT................................................................................................................................................................................ | 20 | 3 | 20 | 3.60 | 72 | 24 | 0 | 0 | 9 | 7 | 5 | 5 | 4 | 3 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| .......................................................................................................................................................................................................................ATTGATAACGTTCTAAATCGT............... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TTCCATGTTCCCTTAAAAGTT.......................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGATGGATACGAATGGCG............................................................................................................................................................................ | 18 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................ATATAGATGGATACGAATGGC............................................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CACAATATAGATGGGTACGA.................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AAATCTGGTCCCGACGTGATCTGTTGTTTTTCGCTACCTGTAACGCGTTCCACGCGTTAACTCTACCCATGCTTACCGTGCTTAAAGCCTCATACATATATAAAAATAAACAAACGCAAAATAAGAAGATTTTATTCTCTTGAATAAATTTAATATATAAAAAAATCTAAAAAAGGTACAAGGGAATTTTCAAATATTTAATACAATATTTTAAGAAACTATAGCAAGATTTAGCACCTTAGTTATAAAAT
**************************************************......((((.....(((.((..((((...((((((.((((.(((((((.((((..((..((((..((((((......))))))....))))..))..))))))))))).)))).))))))...)))))).))).....))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
V047 embryo |
M027 male body |
SRR060682 9x140_0-2h_embryos_total |
SRR060678 9x140_testes_total |
M047 female body |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......GGTCCCGACGTGATCTGTT.................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CTAAAAAAGGTACAAGGGAAT................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CGTGCCACGCCTTATCTCT........................................................................................................................................................................................... | 19 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| .....TGGCCCCGACGTGAACTG.................................................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TTTAAGAAACTTTAGCAA........................ | 18 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................TGTTGTTTTGCGCTAGCAG................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................TTCTTCGCTGCCTGTAACA.............................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................ATATGAGAAAATCTAAAAA.............................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:9570118-9570368 - | dvi_16185 | TTTAGACCAGGGCTGCACTAGACAACAAA---AAGCGATGGACATT-GCGCAAGGTGCGCAATTGAGATGGGTACGAATGGCACGAATTTCGGAGTATGTATATATTTTTATTTGTTTGCGTTTTATTCTTCTAAAATAAGAGAACTTATTTAAATTATATATTTTTTTAGATTTTTTCCATGTTCCCTTAAAAGTTTATAAATTATGTTATAAAATTCTTTGATATCGTTCTAAATCGTGGAATCAATATTTTA |
| droMoj3 | scaffold_6540:7677396-7677470 - | T-GAAACCAGGGCTACGCCAGCTAACAAGTGTTAGGGATGTGCACTAGTCCCTGCTGTACACTTGAGATGGCTGCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14906:2458942-2458981 - | T--GTGCCAGGGCTGCACTAAATACCAAT---TAGATATGAGCGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:19 PM