
ID:dvi_16054 |
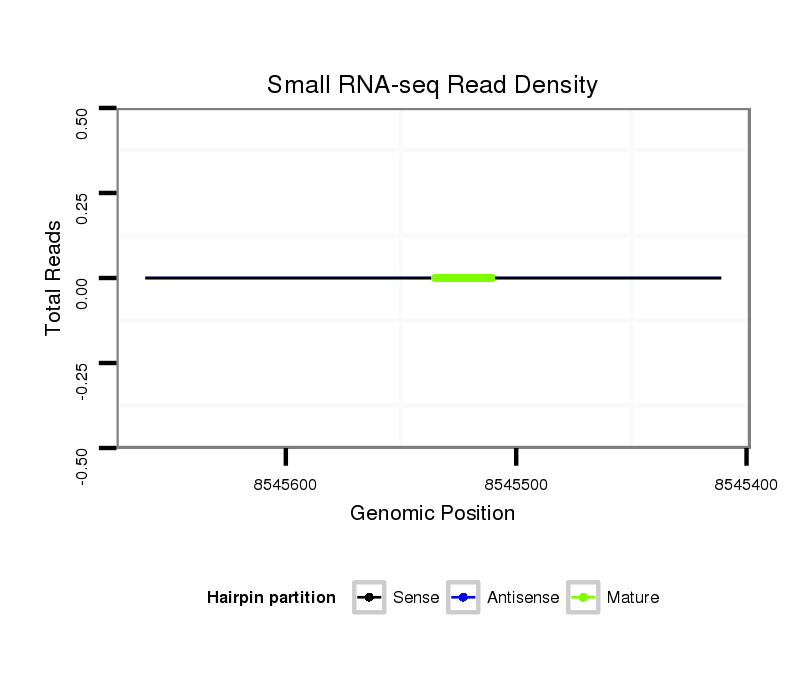
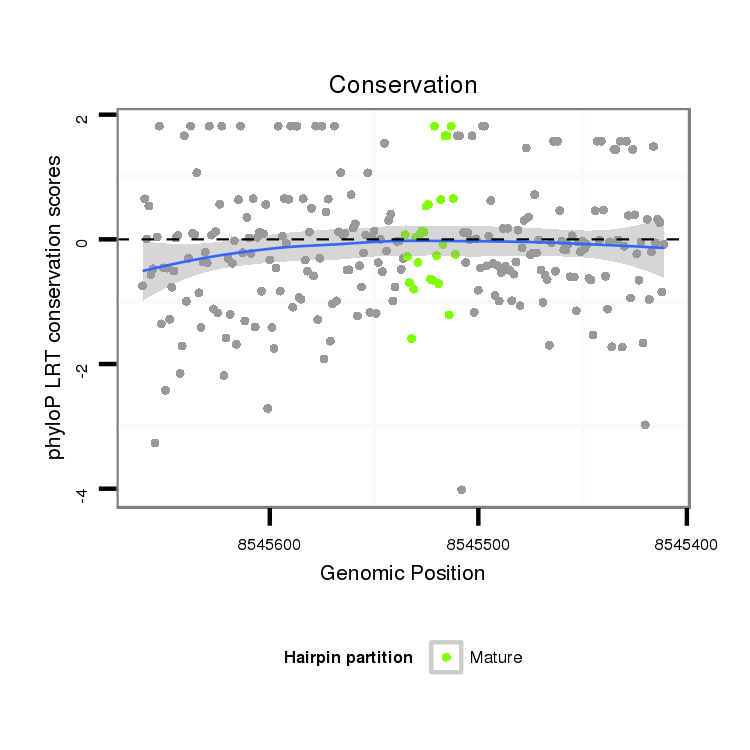
Coordinate:scaffold_13047:8545461-8545611 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.5 | -30.8 | -30.8 |
 |
 |
 |
CDS [Dvir\GJ23148-cds]; exon [dvir_GLEANR_8509:1]; intron [Dvir\GJ23148-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTTGTCGCAAGTTATACCATCCAATCTGGAGGAGTTTCTACGCATCGTCAGTGAGCAAGCGAATGAATACAACTTGGTTAATTTAATTGTGCTGCACTCAACTTTGTACAATTTCGAAGGATCGATTAGTGCATATCGTCTACAGCCGTTTCCGAGTCCGATTTTCGAAAGCGTTTTAGACATAAATGACGGTCCTATATTTCCCAAGGTCTGGCTGAATTTTCGTGGAAAGACAGCTGTTATATTGCCCG **************************************************.(((.((((.............)))).)))....((((((((.............))))))))....(((((((.(((.....(((.(((((.(((.((((.((((.......)))).))))))).))))))))..)))))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060684 140x9_0-2h_embryos_total |
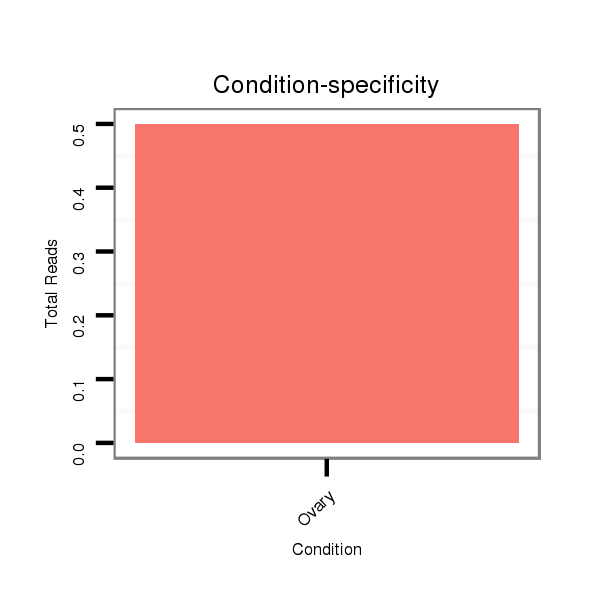
SRR060688 160_ovaries_total |
V116 male body |
SRR060659 Argentina_testes_total |
V047 embryo |
M027 male body |
SRR060666 160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060680 9xArg_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TGTATCTCCTCTACAGCCGTT..................................................................................................... | 21 | 3 | 2 | 2.50 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GCCACTGAATCCAACTTGGTT............................................................................................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AGCCACTGAATCCAACTTGGT............................................................................................................................................................................. | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TAGTGCATATCGTCTACAGCCGTTT.................................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GCCACTGAATCCAACTTGGT............................................................................................................................................................................. | 20 | 3 | 5 | 0.40 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GGCTTAATGTTCGTGGAAA.................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CTGAATCCAACTTGGTTGAT......................................................................................................................................................................... | 20 | 3 | 11 | 0.27 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................................TTTAGAAATAAATGACGGA.......................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGAATCCAACTTGGTTGAT......................................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CACTGAATCCAACTTGGTT............................................................................................................................................................................ | 19 | 3 | 20 | 0.20 | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................GGTGTGGCTGAATTTCCGTT........................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AACCTTGTACATTTGCGAAG.................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................TTACCCAAGTCTGGCTGAA................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTTTTGTAGAATTTCGAAG.................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................ATGATTACAACTTGATGAAT......................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GAACAGCGTTCAATATGGTAGGTTAGACCTCCTCAAAGATGCGTAGCAGTCACTCGTTCGCTTACTTATGTTGAACCAATTAAATTAACACGACGTGAGTTGAAACATGTTAAAGCTTCCTAGCTAATCACGTATAGCAGATGTCGGCAAAGGCTCAGGCTAAAAGCTTTCGCAAAATCTGTATTTACTGCCAGGATATAAAGGGTTCCAGACCGACTTAAAAGCACCTTTCTGTCGACAATATAACGGGC
**************************************************.(((.((((.............)))).)))....((((((((.............))))))))....(((((((.(((.....(((.(((((.(((.((((.((((.......)))).))))))).))))))))..)))))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060661 160x9_0-2h_embryos_total |
V047 embryo |
SRR060664 9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................AGATCCGAAGCAGTGACTC.................................................................................................................................................................................................... | 19 | 3 | 7 | 0.57 | 4 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| ....................................AGATCCGAAGCAGTCACTGG................................................................................................................................................................................................... | 20 | 3 | 4 | 0.50 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGATCCGAAGCAGTGACTCG................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGATCCGAAGCAGTGACT..................................................................................................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AAGATCCGAAGCAGTGACT..................................................................................................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TTCACGTACAGCAGATTTCGG........................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................AGGTTAGATCTCCTCATGG..................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................CTATCGACAATATAACGA.. | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................CAAGCTTCCTAGCTACTCG......................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................TGGGTTCGAGACGGACTTA............................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................ATCCGAAGCAGTCACTGG................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:8545411-8545661 - | dvi_16054 | CTTGTCGCAAGTTATACCATCCAAT---CTGGAGGAGTTTCTACGCATCGTCAGTGAGCAAGCGAATGAATACAACTTGGTTAATTTAATTGTGCTG---------CACTCAACTTTGT----------------ACAATTTCGAAGGATCGATTAGTGCATATCGTCTACAGCCGTTTCCGAGTCCGATTTTCGAAAG------------CGTTTTAGACATAAATGACGGTCCTATATTTCCCAAGGTCT-GGCTGAATTTTCGTGGAAAGACAGCTGTTATATTGCCCG |
| droMoj3 | scaffold_6540:27614287-27614537 + | TCTGCTGCAGACTAAGTCATCCAAT---CTTCAAGATGATTTCGCTATAATCGGTGAACAGGCCAATAATTACAATTTTGTCAGTCTGATCGTGTTG---------CACTCTACTTCAT----------------TCGATGACAAAGAGCCGATTGTCGCATATCGTCTTCAGCCGTTCCCCAGTCCAAGTTTTGCAACTATTAGCA---ATA---------TAAATAATGAACAGATTTTTCCGAAAGTGG-GGCGTGACTTCAAAGGAAAAACGGCTCTCATTTTACCCG | |
| droGri2 | scaffold_15074:1070360-1070610 + | CTTGTTAAATAGTGTGGCGTCAAAT---CTAACAAAGGACCTGCAAATAATTTGTGAAACTACTGCGAACTATAACTTCATCCATTTAATTGTTCTA---------CACTTAACGTCGA----------------ACCGTATCGAAGAATCGATTGTAACATATCGATTGCAACCATTTCCGAGTCCAACTTTAGAGCGCATCCGAA---ATA---------TAAGTGACGGACACATATTTCCCAAGATGT-GGTTAAATTTTAAGGGGAAAACGGCATTGTTTTTACCAG | |
| droWil2 | scf2_1100000004943:9550979-9551144 - | CTGGATTAATATTCCGCATCCTAACGAGTTGTCGCAGCTTCTTTTCATAATTGGCAACCAAGCAACTAAGCATAAATTCATCAATTTATTGGCTATA---------CATCAG----------------------------ACAAACGACACGGTCATTGGATTTCACTTGAAACCTTTCCCTGTAGCGCATTTTGAAAGAGTT----------------------------------------------------------------------------------------- | |
| dp5 | 2:12108405-12108643 - | GTGGACGCAAGGGAATCCCACAGA---------GGAATTCCTCAAACAGATCTCCGACAAGGCCGAGGAGCATAAATTTCTTCATATGATCGTTCTGGGA-------------CTCCATCAAAGTGG------------------------GGAAACGATGCATCGATTGAAACCATTTCCCAGTCCCCGCTTTGAGAGGATCGAGA---ATGTTTCGTGCATAAAG---GGATCCGTTTTCGACC-GGCCTACACTCAACTTCCAAGGAAGAACGGCGACCGTGCTGCAAA | |
| droPer2 | scaffold_0:8388234-8388472 + | GTGGACCCAAGGGAATCCAGCAGA---------GGAATTCCTCAAACAGATCTCCGACAAGGCCGAGGAGCATAAATTTCTTCGTATGATCGTTCTGGGA-------------CTTCATCAAAGTGT------------------------GGAAACGATGCATCGATTGAAACCATTTCCAAGTCCCCGCTTTGAGAGGATCGAGA---CTGTTTCGTTCATAAAG---GAATCCGTTTTCGCCC-GGCCTACACTCAACTTCCAAGGAAAAACGGCGACCCTGCTGAAAA | |
| droEle1 | scf7180000490717:7752-7981 + | TTTTATGCAGATCTTGCCTTCTCA---------GAAGTTTCTCGAAGACATCGCACATCAGGCAGCAAAGCACAAGTTCCTCAATCTCGTGGTAATC----------------------------------------------GAAAAGTCCAAAAAAATCTATAGATTGTTTCCATTTCCCCAGCCAAAGTTCGAAGTGTTCCGCTTCAAACCGCTTGAAGGA------AAGGAAATCTTTCCCGCTTCCT-GGAACAACTTTATGGGAAAGATCGCTAAAACAATGCCCG | |
| droBia1 | scf7180000302402:5797224-5797448 + | GTGGAGCCAAAGAGAACCCTCTAA---------GGAGTTACTAGATCACATCTCCAGGCTGTCCATCAAGTTTAAGTTTGCTCAAATACTTATGCTGGCA-------------ATGGCCGATAATACCGAAGGAA---------------ATCCAACATTGTACCGAATGAATCCGTACCCGACTCCTCAATTTAAAAA------------TGTATCTAACATCAAA---AGTCCAGTTTTCCTAG-AATCTGGCTACAACTACTATGGTATGACGGC-------------- | |
| droTak1 | scf7180000415711:1124614-1124860 + | GTGGCTTCAGGCTAAACTCACCCA---------AGAGATGCTTAATGTGATTGCCAATCAAGTGGAGGAACATCAATTTCTTCAAATGCTCATCTTAGAGGTTAGAG----------------------------ACGAACCAAAGGAAATGGTAGCTGTACGACGTCTTCATCCTTTTCCTACTGCACATTTTCACCGAATCGATA---GCGTATTCGACCACAAG---GGAGCCATTTCCATCC-GCCCG-AGATGAACTTTAAAGGAAGGACCGTTAATCTGCTGCCTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/19/2015 at 07:49 PM