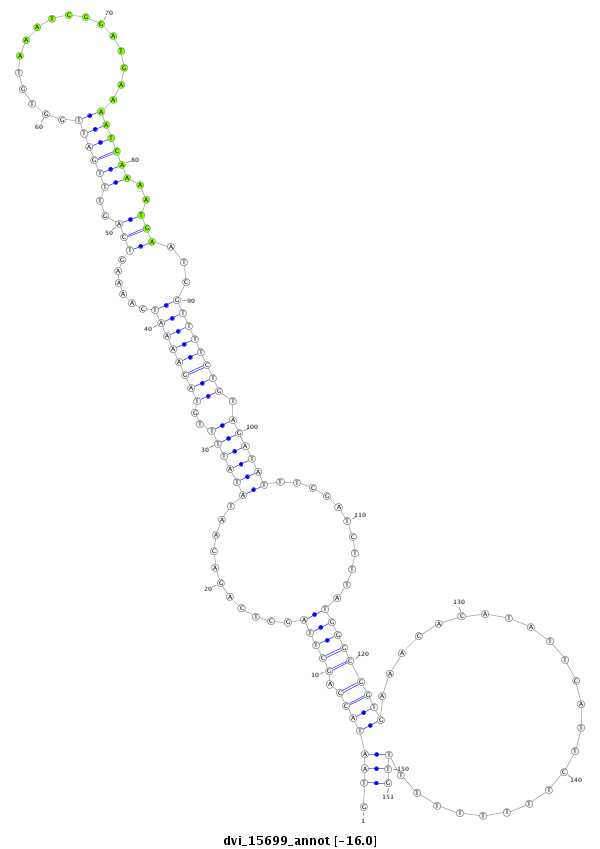
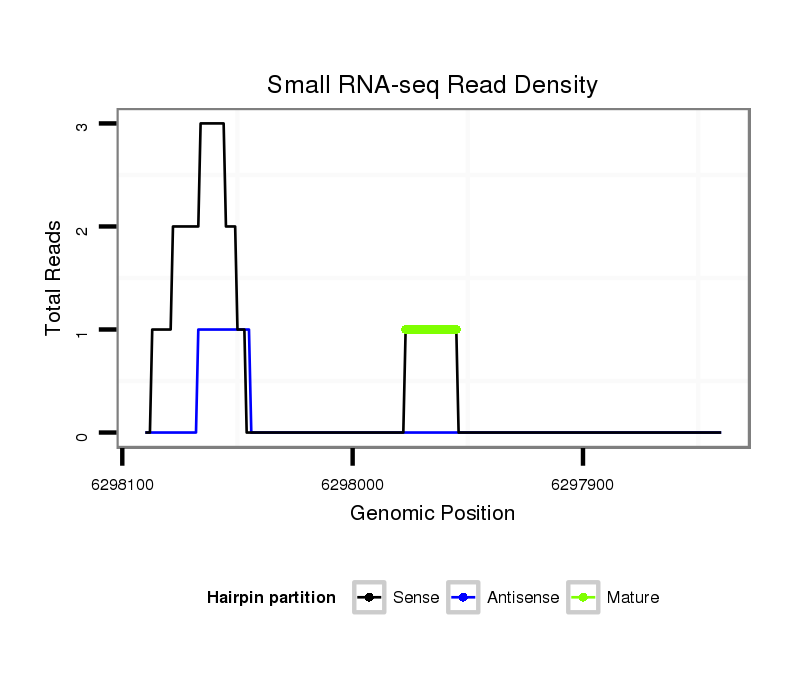
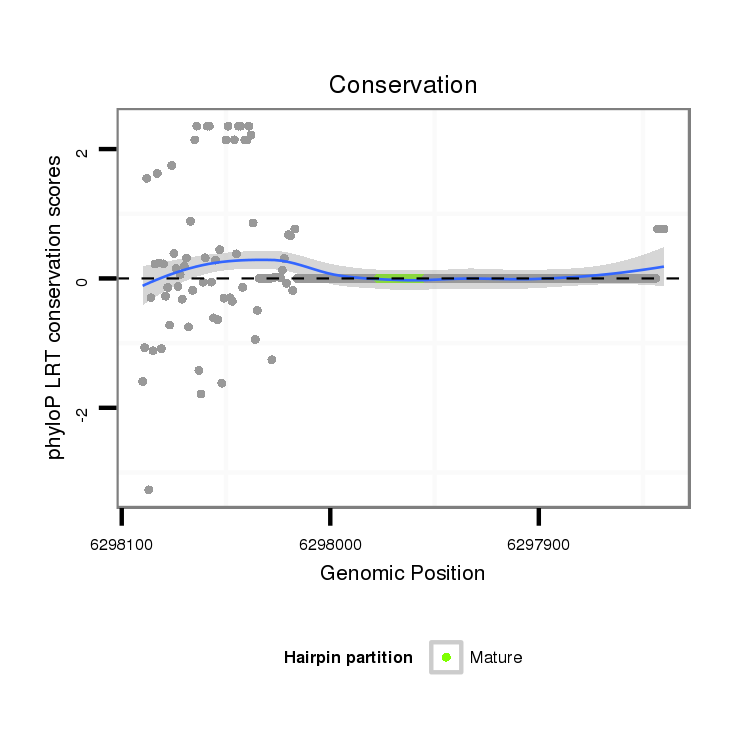
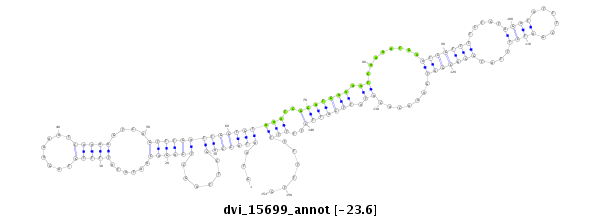
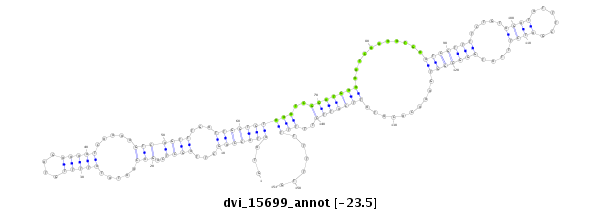
ID:dvi_15699 |
Coordinate:scaffold_13047:6297890-6298040 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.0 | -23.6 | -23.6 | -23.5 |
 |
 |
 |
 |
CDS [Dvir\Taf1-cds]; exon [dvir_GLEANR_8646:3]; intron [Dvir\Taf1-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CACTGCTGATGGAACTAGTTCCACCGACGAGAAGTCAAGTGAGGCCAAAGGTAATACCAGCTTAGCTCAGACAATATATTTTGTAGAAAATCAAAAGTCAGTTTGATTGGTGTAAATCGGATGAAAATCAAAATGAATCGTTTTCTGTAGATATTTCGATCTTTATGGGCGGTGAAACACATATTCATTCTTTTTTTTTTGTGTATAAAGCTGTGGAAATAAGTTTGAAGCCGATCGTATGAAAATTGAAC **************************************************.(((((((.(((((...........((((((..((((((((......(((..((((((.................))))))..)))...)))))))).))))))...........)))))))))........................)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060686 Argx9_0-2h_embryos_total |
M061 embryo |
SRR060657 140_testes_total |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
V047 embryo |
SRR060685 9xArg_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................CACCGACGAGAAGTCAAGT................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................CGACGAGAAGTCAAGTGAGG............................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGCTCCACCGACGAGAAGTCAAGTGA................................................................................................................................................................................................................. | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...TGCTGATGGAACTAGTTC...................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................AAATCGGATGAAAATCAAAATGA................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AACTAGTTCCACCGACGAGAAGT........................................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ATAAGTTTGAAGCCGATCGT............. | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GAAGAAAATCAAGATGAATTGTT............................................................................................................. | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................AGTCAGATTGATGGGTGT.......................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................TTTAAGGGCGGTGAAGCA........................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................ATATATCGATCTTTGCGGGC................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTGACGACTACCTTGATCAAGGTGGCTGCTCTTCAGTTCACTCCGGTTTCCATTATGGTCGAATCGAGTCTGTTATATAAAACATCTTTTAGTTTTCAGTCAAACTAACCACATTTAGCCTACTTTTAGTTTTACTTAGCAAAAGACATCTATAAAGCTAGAAATACCCGCCACTTTGTGTATAAGTAAGAAAAAAAAAACACATATTTCGACACCTTTATTCAAACTTCGGCTAGCATACTTTTAACTTG
**************************************************.(((((((.(((((...........((((((..((((((((......(((..((((((.................))))))..)))...)))))))).))))))...........)))))))))........................)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
|---|---|---|---|---|---|---|---|
| .......................GGCTGCTCTTCAGTTCACTCCGG............................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .TGACGACTTCCTTGGTCAAGG..................................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .....GACTTCCTTGGTCAAGG..................................................................................................................................................................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 |
| ...........................................CGGTTTCCACTCTGGTCGG............................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:6297840-6298090 - | dvi_15699 | CACTGCTGATGGAACTAGTT---CCA---CCGACGAGAAGTCAAGT---GAGGCCAAAGGTAATACCAGCTTAGCTCAGACA----------ATATATTTTGTAGAAAATCAAAAGTCAGTTTGATTGGTGTAAATCGGATGAAAATCAAAATGAATCGTTTTCTGTAGATATTTCGATCTTTATGGGCGGTGAAACACATATTCATTCTTTTTTTTTTGTGTATAAAGCTGTGGAAATAAGTTTGAAGCCGATCGTATGAAAATTGAAC------------ |
| droMoj3 | scaffold_6540:1777521-1777585 + | ACCAGCTGAAGCGTCTG------GCGGCAGCGACGAAAAGTCAAGC---GACGCTAAAGGTAGCT------GAGCTTAG--A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14906:11618178-11618223 - | CACCAGCGATGCA------------TCAAGCGAATCCAAATTGAGT---GACGCCAAAGGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004902:11791316-11791366 + | GACCACT------------G---CTAGTGGTGAAACCAAAAGTCAG---GATGCCAAAGGTAATT------C-----AA--G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | 2:19899944-19900006 - | CCCTGATGGCGGCGCTGCATCAGCCTGTAGTGAGACCAAGGCCAGTGGGGACCCCAAGGGTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_3:2648490-2648561 - | CCCCGATGGCGGCGCTGCATCAGCCAGTAGTGAGCCCAAGGCCAATGGGGACCCCAAGGGTAAGC------G----CAAACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13340:5560083-5560124 - | CACTA------------------CCACAAGCGATAGCAAGGCCATC---GATCCCAAAGGTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396708:3253775-3253832 + | AACGACTGAAACCTCTACAA---CCAGTAGCGATAACAAGGCCATC---GACACCAAGGGTAAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302619:189784-189838 + | TT--GAAGCCACCACCAGTC---CCAGTGGCGACGCCAAGACCAAT---GATGCCAAAGGTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302155:2207042-2207091 - | CG------AGGGCTCCAGTC---CCGGCGGCGACACCAAGTCCTCC---GATGCCAAAGGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409791:309832-309923 + | TG------ATGCCACCAGTCCAAGTGCTAGTGATTCCAAAGCAAAC---GATCCCAAGGGTAAGC------ATATACCG--CAATACAGCTAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAACAATACTTTATGT | |
| droYak3 | 3R:7943611-7943661 - | CG------ATGCAACCAGTC---CCAGTGGTGACCCCAAATCTATC---GATGCAAAGGGTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4770:2715243-2715298 + | TC-AAGTGATGCAACCAGTC---CCAGTGGCGATCCCAAATCTACC---GATGCAAAGGGTAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:59 PM