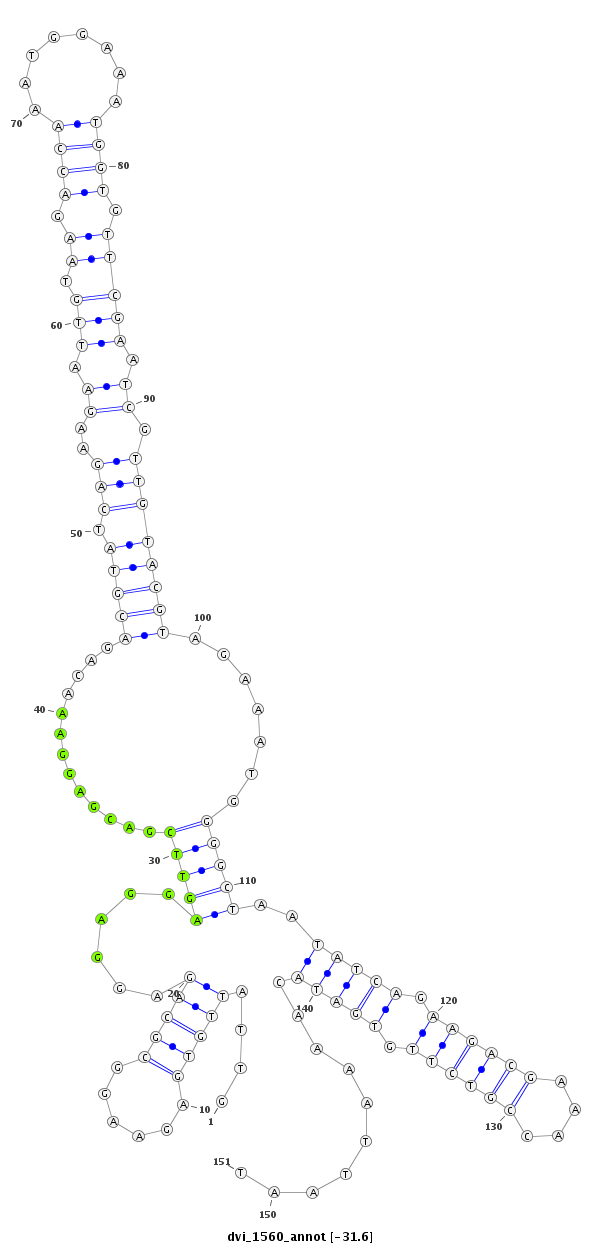
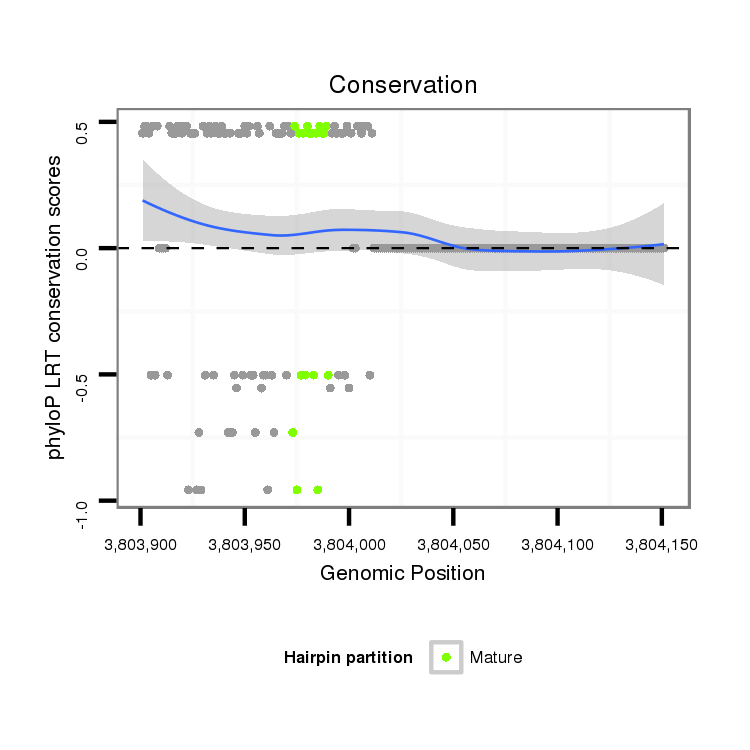
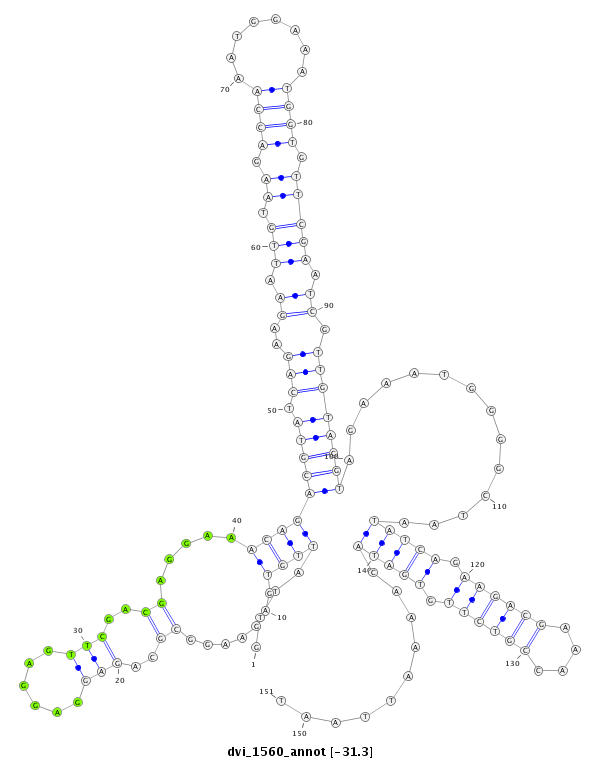
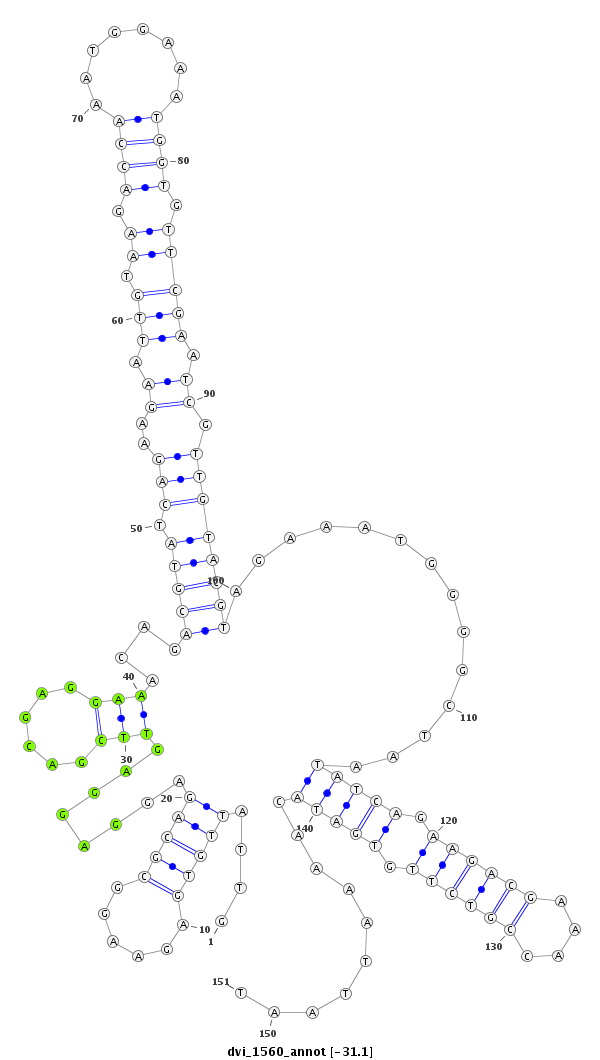
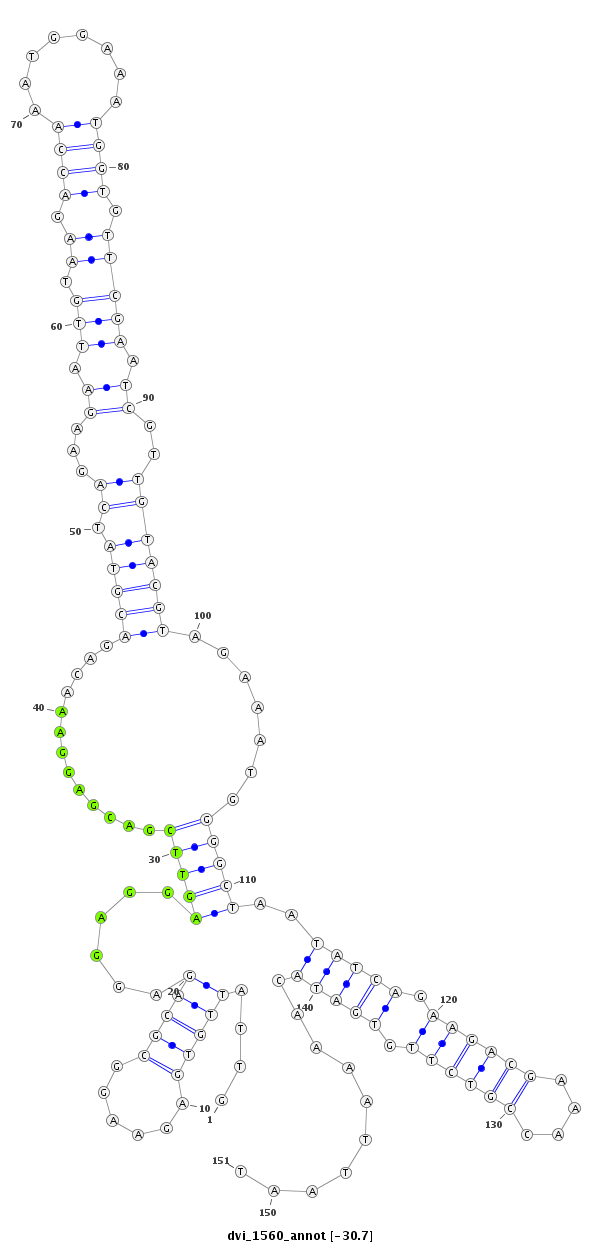
ID:dvi_1560 |
Coordinate:scaffold_12723:3803951-3804101 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.3 | -31.1 | -30.7 |
 |
 |
 |
CDS [Dvir\GJ18295-cds]; exon [dvir_GLEANR_2852:1]; intron [Dvir\GJ18295-in]; Antisense to intron [Dvir\GJ17939-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATCGACATCAGGTGGCAACAAGGGCGGCAAGAGAAGGAGGCGCGTCCGAGTTATTGTGAGAAGGCGCAGAGGAGGAGTTCGACGAGGAAACAGACGTATCAGAAGAATTGTAAGACCAAATGGAAATGGTGTTCGAATCGTTGTACGTAGAAATGGGGCTAATATCAGAAGACGAAACCGTCTTGTGATACAAAATTAATTGCTGATAGCCTCTAAGCCTCAATAAAATTAAATTGCATAAAAGAAAGAA **************************************************....(((((......)))))......(((((.............(((((.(((..((.(((.((.((((........)))).))))).)).)))))))).......)))))..(((((.((((((....)))))).)))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
M027 male body |
SRR060655 9x160_testes_total |
SRR060664 9_males_carcasses_total |
M028 head |
SRR060677 Argx9_ovaries_total |
V053 head |
SRR060689 160x9_testes_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................TAATCGCTGAAAGCATCTAAGC................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTATCTCAGAAGATGAAACCG...................................................................... | 21 | 3 | 7 | 0.71 | 5 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................GAGGAGTTCGACGAGGAA................................................................................................................................................................. | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GCTGAACGCCTCTAAGCC............................... | 18 | 2 | 5 | 0.40 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................ATTGCTGAACGCCTCTAAGTC............................... | 21 | 3 | 5 | 0.40 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TTAAGACCAAATGGAAATG.......................................................................................................................... | 19 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GCTGAAAGCCTCTAAGTC............................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................ATTGCTGAACGCCTCTAAG................................. | 19 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................ACAATGGCGGTAACAGAAGG..................................................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................GTCGAGGAAACAGGCGTTT....................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................GCTGATCGCCTCTAAGTCA.............................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CAAATGGAAAAGGAGTCCGA.................................................................................................................. | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................CGCGTCATCGTTATTGTGA............................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GTAGCTGTAGTCCACCGTTGTTCCCGCCGTTCTCTTCCTCCGCGCAGGCTCAATAACACTCTTCCGCGTCTCCTCCTCAAGCTGCTCCTTTGTCTGCATAGTCTTCTTAACATTCTGGTTTACCTTTACCACAAGCTTAGCAACATGCATCTTTACCCCGATTATAGTCTTCTGCTTTGGCAGAACACTATGTTTTAATTAACGACTATCGGAGATTCGGAGTTATTTTAATTTAACGTATTTTCTTTCTT
**************************************************....(((((......)))))......(((((.............(((((.(((..((.(((.((.((((........)))).))))).)).)))))))).......)))))..(((((.((((((....)))))).)))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12723:3803901-3804151 + | dvi_1560 | CATCGACATCAGGTGGCAACAAGGGCGGCAAGAGAAGGAGGC-GCGTCCGAGTTATTGTGAG-AAGGCGCAGAGGA------------GGAGTTCGACGAGGAAACAGACGTATCAGAAGAATTGTAAGACCAAATGGAAATGGTGTTCGAATCGTTGTACGTAGAAATGGGGCTAATATCAGAAGACGAAACCGTCTTGTGATACAAAATTAATTGCTGATAGCCTCTAAGCCTCAATAAAATTAAATTGCATAAAAGAAAGAA |
| droMoj3 | scaffold_6500:14969103-14969221 - | CATCAATA----ATGGCAACAACGGCCTGAGGAGGAGGAGGAAGAAACCAAGTCGGTGAAGCGAGTGCGCAAAGTAAAAAAAAGAACTCGGGCTCGGCCAGGAGTCAGGCGCAAC--AAGAATCG-------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/19/2015 at 11:23 AM