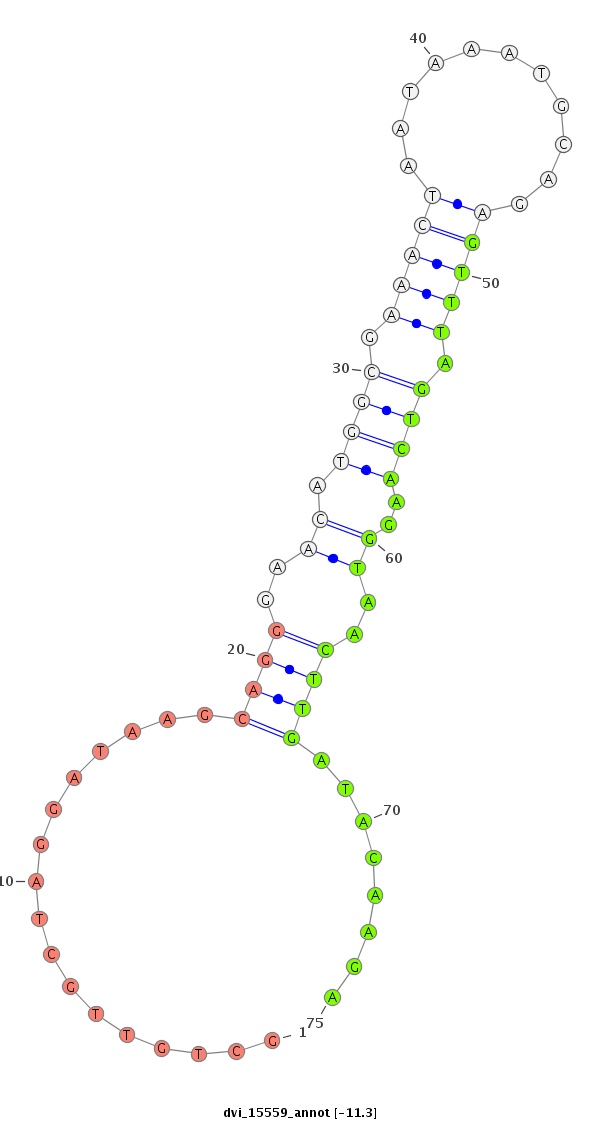
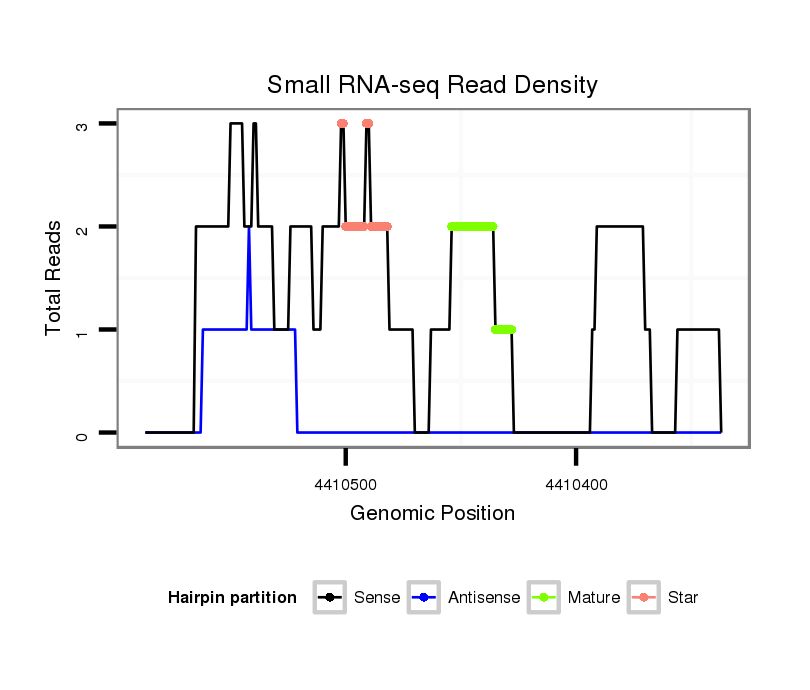
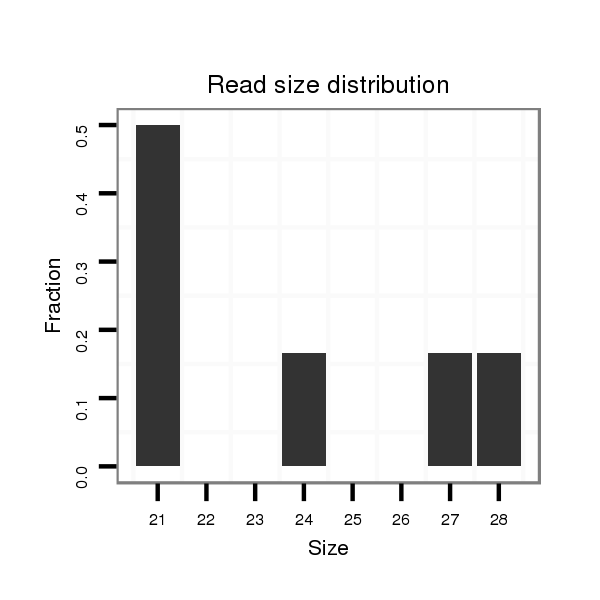
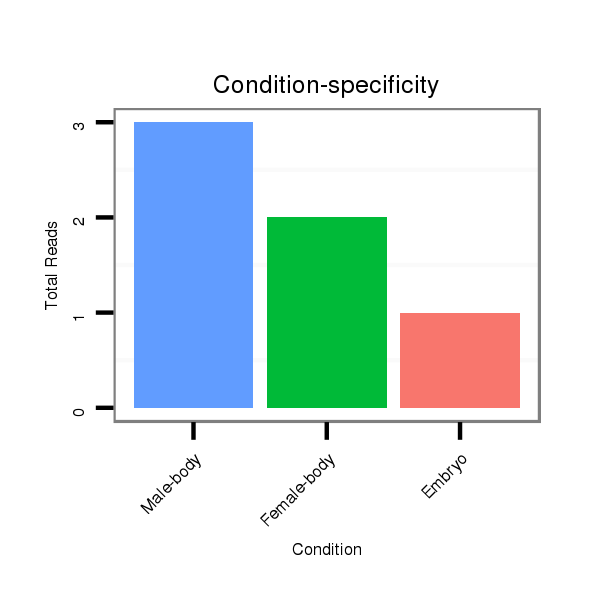
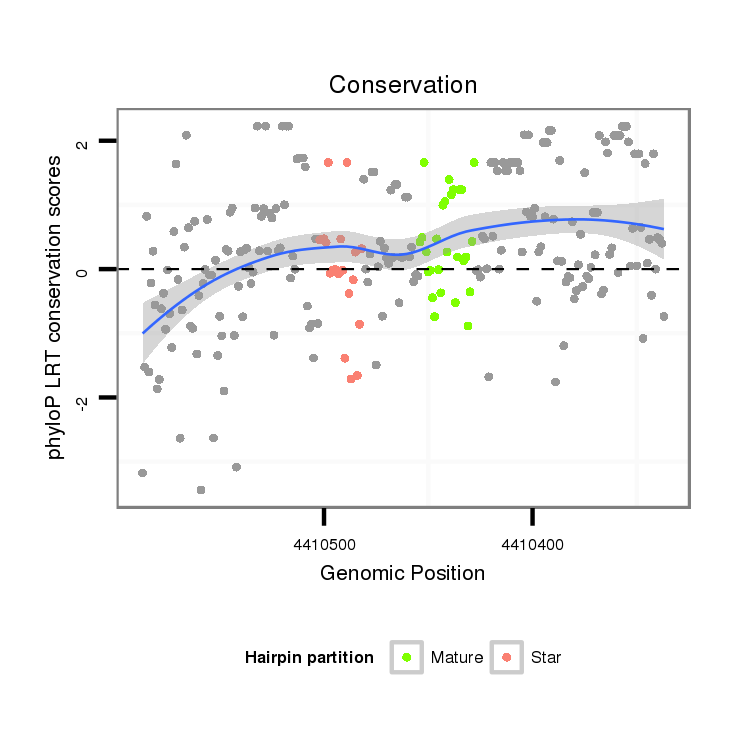
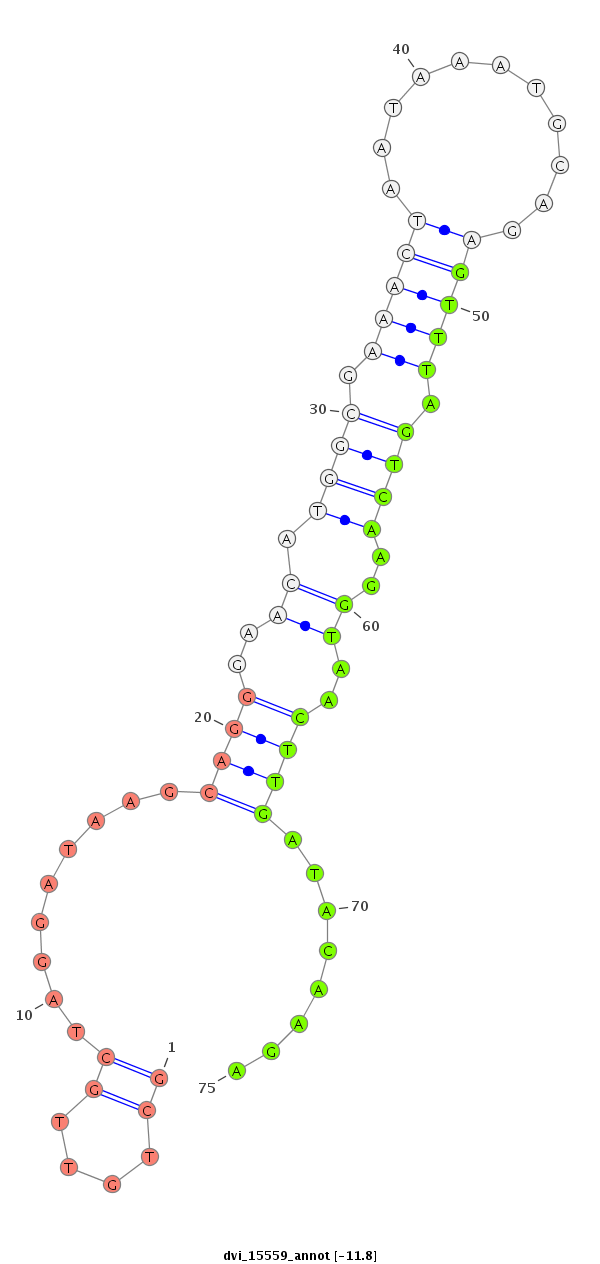
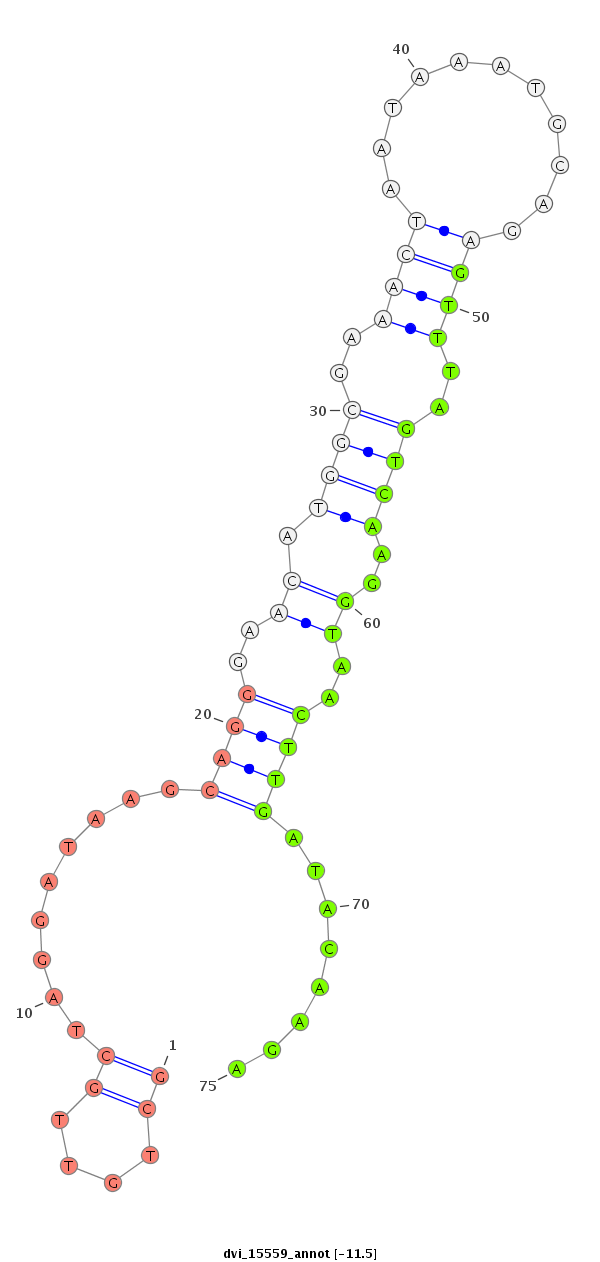
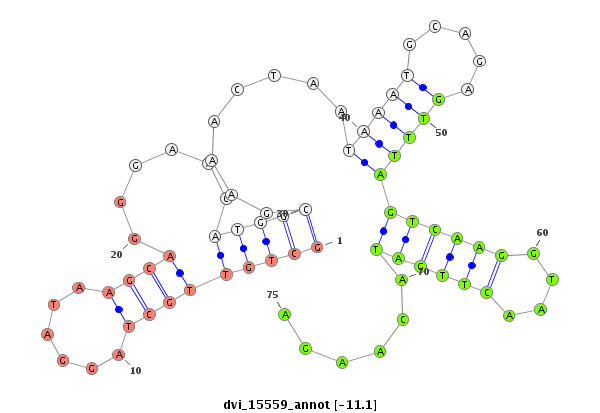
ID:dvi_15559 |
Coordinate:scaffold_13047:4410387-4410537 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -11.8 | -11.5 | -11.1 |
 |
 |
 |
exon [dvir_GLEANR_8715:3]; CDS [Dvir\GJ23358-cds]; intron [Dvir\GJ23358-in]
| Name | Class | Family | Strand |
| Helitron-2N1_DVir | RC | Helitron | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#######################--------------------------- TCTTGATGAAAACAGAATAAATTGCCAGACAAGAACCTGCTCTTCGACAAAATGTTCCTACGGTAACTATATGATATAGAGGTCTGCTGTTGCTAGGATAAGCAGGGAACATGGCGAAACTAATAAATGCAGAGTTTAGTCAAGGTAACTTGATACAAGAACCTACCTATGACAGCTATATGATATAGTTGTCCGATTTAGTTGGTTTCGGCAAGCTGATCTAGAATATATATATTTCATCGATCTAGGCT *************************************************************************************.................((((..((.((((.(((((...........))))).))))..))..))))........******************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M061 embryo |
M047 female body |
M027 male body |
SRR060676 9xArg_ovaries_total |
V053 head |
V047 embryo |
GSM1528803 follicle cells |
SRR060662 9x160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................ATATTTCATCGATCTAGGC. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................GATTTAGTTGGTTTCGGCAAGCTGAT............................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTTAGTTGGTTTCGGCAAGCT.................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TGCCAGACAAGAACCTGCTCTTCGACA.......................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GTTTAGTCAAGGTAACTTGATACAAGA........................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGTAGGATAAGCAGGGAACATGGC........................................................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................ATTGCCAGACAAGAGCCTGCT.................................................................................................................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AAATGCAGAGTTTAGTCAAGGTAACTTG................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TGCTCTTCGACAAAATGTT................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GTAGAGGTCTGCTGTTGCTAGG.......................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................GCTGTTGCTAGGATAAGCAGG................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................AGAGGTCTGCTGTTGCTAGGA......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TGCCAGACAAGAACCTGCTCT................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GATAAGCAGGGAACATGGCGA...................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TAACTATATGATATAGAGGTCTGC.................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CAAAATGTTCCTACGGTAACTATATG.................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........AACAGATTAAATTGACAGAC............................................................................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GATTCGGCAAGCTGAT............................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GCAACTTGATCTAGAAGATAT.................... | 21 | 3 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ..................................................................................................TAAGCAGGAAAAATGGCG....................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AGAACTACTTTTGTCTTATTTAACGGTCTGTTCTTGGACGAGAAGCTGTTTTACAAGGATGCCATTGATATACTATATCTCCAGACGACAACGATCCTATTCGTCCCTTGTACCGCTTTGATTATTTACGTCTCAAATCAGTTCCATTGAACTATGTTCTTGGATGGATACTGTCGATATACTATATCAACAGGCTAAATCAACCAAAGCCGTTCGACTAGATCTTATATATATAAAGTAGCTAGATCCGA
*******************************************************************************************.................((((..((.((((.(((((...........))))).))))..))..))))........************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060680 9xArg_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060654 160x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................GTCTGTTCTTGGACGAGAAGC............................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTCGTTGGATACTGTCGATATACTAT.................................................................. | 26 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TGCCATTGGTATACTATATCT........................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CTGTTTTACAAGGATGCCATT......................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TCGTTGGATACTGTCGATATACTAT.................................................................. | 25 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................GTTCTTGGACATGAAGCT............................................................................................................................................................................................................ | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................GGATACTGTCGATATACTATATCAA............................................................. | 25 | 0 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................TGTTTAACGGTCTGTTGTT........................................................................................................................................................................................................................ | 19 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................TTCAACTAGATATTATATAT................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................ACTGTCGATATACTATATCAACAGGT........................................................ | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:4410337-4410587 - | dvi_15559 | TCTTGATGAAAACA------------------------------GAATAAATTGCCAGACAAGAACC----TGCTCTTCGACAAA-ATGTTCCTACGGTAACTATATGATATAGAGGTCTG-CTGTTGCTAGGATAAGCAGGGAACATGG-CGA----------------------------AACTAATA---AAT-------------GCAGAGTTTAGTCAAG----------------GT---------------------AACTTGATACA----AGAACCTACCTATGACAGCTATATGATATAGTTGTCCGATTTAGTTGGTTT----C-------GGC-------------------------------------------------------------------------------------------------------------------------------------AAGCTGAT------------------CTAGAATA-----------------TATATATTTCATCGATCTAGGCT--- |
| droGri2 | scaffold_15074:4012488-4012741 + | TCTTGATTTGG-----------------ATAACA-AGCTAAACACAAAAGTTGCTCATACGAAAGCT----TATCTTTCGACTGT-TTTTTTCAACGTTTGCTATATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATGGTTATCCGATCTAGCCGGCTC----A-------GATATAT---------GCAGC--AAAAAGACGCATATATTCCAAGTTTCATCAAGAGCGCTTCAAGACTGAGAGACAAGTTGTTATAGAAACAGATA--------------------------GACTCTGTTAGTGATGCTGAT------------------CAAGAGTATA---------------TATATACTTTATGGAG-TCAGAGACT | |
| droWil2 | scf2_1100000004516:2192226-2192492 + | GCTTCAAAA-------------------GTAACGAAG----------TTA-TTGAC---AAAAGTCA----CTGTTTTCAACCGA-TCGTTCCTATGGGAGCTATATGATATAGTAACCCGATCTTTATCAAATTCAAAACAG-TCATTAACAGATAT-----------ATTA---------AACTAACA---AAT-------------GTTTAATTTGAAAGTTATTGACAAAA------GTC---ACTGTTT--------------------TCGAAAGATCGTTCCTATGGGAGCTATATGATATAGTCACCCGATCT-----------------------------------------------------------------------------------------------------------------------------------------------------------------TGATCAAATTTGGCATAGTCGTTT---ATATGTGTAATTAACTCAACAATAT-----------------T--- | |
| droAna3 | scaffold_9723:12513-12743 - | TTTATTTGCAG-----------------ACAACG----TG----AAACGG-TTGACAGTCA-------------TTTGCAA--GCTCTGGTTATATGGCAGCTATAAGATATAGCCGAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAG--GAAAAGAAGGATGTGTGCAAAGTTTCAACTAGATAGCTTTAAAACTGAGAGACTAGTTAGCGTAGAAACAGACAGACGGGCAGACCGATAT-GCTATATCGACAAAGGAGGTGATTCTGAT------------------CAAGAATA-----------------TATATACTTTATAGGG-TCGGAG--- | |
| droBip1 | scf7180000391788:2782-3040 + | AA--------GGCG------------------------------AAATAGTT-TCCCGGCAAAAACACTAATCGTTTTACACTGA-ATGTTTCTATGGCAGCTATAACCTATAGTATCCAGATTTTTACTGAAGTTTCCCAGG-TTATATACGAAAAT--------------------------------------------TTTCAAAGTATTGTTGCATCAAAA-TGAAGAACAATAA-----------ATTTTCATATAAAAACTGAATATGAGATCGATTGCTTATGTGGCAGCTATATGATATAGTTGTCCGATCTGGCAGATTT----T-------AACATA-------------------------------------------------------------------------------------------------------------------------------------TTAAT------------------TTAGGATA-----------------AATATA-------------------- | |
| droKik1 | scf7180000302486:2433039-2433334 + | CGTTGAAAA-------------------ACAACGAATATA----TAATTT-TTTTCTATTA-------------A--TTTCCCGA-TCGTTCCTATGGCAGCTATATGATATAGTCGTCCGATTTTCATAAAATTTA------------TACCAAAATTCAGAAATAATATAAAATGGCCATATCTAAAA---A------------------ATGGTGCAAAAATGTTGAAAAACAGCAAAGTTATAATTTTTT--TCT--AAAAA--------TATATCGAACATTTGTATGGCAGCTATATGATATAGTCGTCCGATCCGGCCCGTTT----C-------GACATATATAGCAGTGAGAGC--ATATAGA--------------------------------------------------------------------------------------------------------------------------------------AGAC---------------------TATATATCATTGAGATAG------ | |
| droFic1 | scf7180000454039:384130-384378 + | GA-------------------------------------------AATAGTT-TCCCGGCGAAAACT----CG-TTTTACACTGA-TTGTTCCTATGGCAGTTATAACCTATAGTATCAAGATTTTTACTGAAGTTTCCCAGG-TTATATACGAAAAT--------------------------------------------TTCCAAATTATTGTTGCTTCAAAA-TGAAAAACAATAG-----------TTTTTCATAAAAAAACTGAATATCAGACTGATTGTTCATATGGCAGCTATATGATATAGTTGCCCGATCTGGCTGATTT----T-------TACATA-------------------------------------------------------------------------------------------------------------------------------------TTATT------------------TTAGGATA-----------------AATATA-------------------- | |
| droEle1 | scf7180000490420:41253-41519 - | TCTAGAAAA-ACTAAAAAAAATTAAAAAACACC---------------------------AAATTTA----ATTTTTTTGTCCGA-TTGTTCCTATGGTAGTTGTATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTATAGTCGTCCGATCCGGCTCGATC----C-------GACTTATATACTAC---CAAT--ATAAAAACAACTTTTGGGAAAGTTTCATGCAGATATCTTTATAACTGAGAGACTAGTTTGCGTAGAAACGGACGG----ACAGACGGACATGGCTAGATCGACTCTCCTAGTGATGCTGAT------------------CAAGAATA-----------------TATATACTTTACAGGG-TCGGAG--- | |
| droRho1 | scf7180000776814:2312-2546 + | GTTTGATGTCAATT------------------------------GC-----TTTTATA----CTGCT----ATGTCATATAACAA-TTTTTCCTACGGAAGCTATA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GACGTCCGATCGG-CAAGCGCTACCCTAATCAAGATA-----------TGCG-TAAATAAATAAGA--CTTGGCAAAATTTATGTCA-GTAGCT--------TACACAC------ACGGACG----GACAGACATACAGACGGACATGGCTATATCGAATCCTCTATTGATCCTGAT------------------CAAGAATA-----------------TATATACTTTATGGGG-TCGGAA--- | |
| droBia1 | scf7180000302098:119627-119875 - | TTTGGATGG-G-----------------ACATCG-------------AAATTTATCAAATGAGGTTT--------TTTTCCCCGA-TAGTTCCTATGGGAGCTATAAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGTAATTGTCTGATCCGGCTGGTTC----C-------GACTTATATACTACTTGCAAT--AGAAAAAAGAAAGTTGGGAAAGTTTCAGCCCGATAGC--------TGATAGACTAGTTTGCGTAGAAACGGACAG----GCAGACGGACATGGCT------ACTCGTCTTGTGACGCTGAT------------------CAAGAATA-----------------TATATACTTTATGGGG-TCGAAA--- | |
| droTak1 | scf7180000414441:10151-10404 + | TTTTAAAAA-------------------ACACCGAAG---------------------CTAGAATTA----AGTTTTTTTTCCGA-TCCTTCCTATGGGAGCTATAAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATAGTTGTCCGATCCGGTCGGTTC----C-------GATATATATACTACCTGCAAT--AGAAAGAAGACTTTTGCGAAAGTTTCGTCCCGATAGCTCAAAAACTTAGAGACTAGTTTGCGTAGAAACAGA--------CAGACGGACATGGCTAGATCGACTCGTCTTGTGATGCCGAT------------------CAAGAATA-----------------TATATACTTTATGGGG-TCGGAA--- | |
| droEug1 | scf7180000407790:2071-2105 - | C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAT------------------TAAGAATA-----------------TATATACTTTATGGGG-TCGGAA--- | |
| droSim2 | 3l:20463742-20463961 + | AACA----------------------------------------AAATGATTTTTCAAACAAAAGCA----CGATTTTCGAATGA-TTGTTCCTATAGCAGCTATATGA-ATAGCATAC-AATTTTCGCTAAGCTCGCCGTGG-ATATGTACA-------------------------------CTAATTTCTTATGTAGAATTCCAAGTTTTCGTTGCAAAATAATTGAAAA-------------------------------CACTTAATTTCAAACTGATCGTTCATATGACAGCTATATGATATAGTGGTCCGATCTGGCTGATTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_101:68057-68304 + | TATTGTTTGCAACA------------------------------AAATCATTTTTCAATCAAAATCA----CGATTTTCGACTGA-TTGTTCCTATAGCACCTATATGATATAGCATAC-AATTTTCGCTAAGCTCACCCTGG-ATATATATA-----------------------------TACCAATTTCTTATGTAGAATTCCAAGTTTTCGTTGCAAAATAATTGAAAAAAAAAAG-----------TTTTCCATACTAACACTTAATTT-----TAATCGTTCGTATGACAGCTATATGATATAGTGGTCCGATCAAGCTGATTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/19/2015 at 07:16 PM