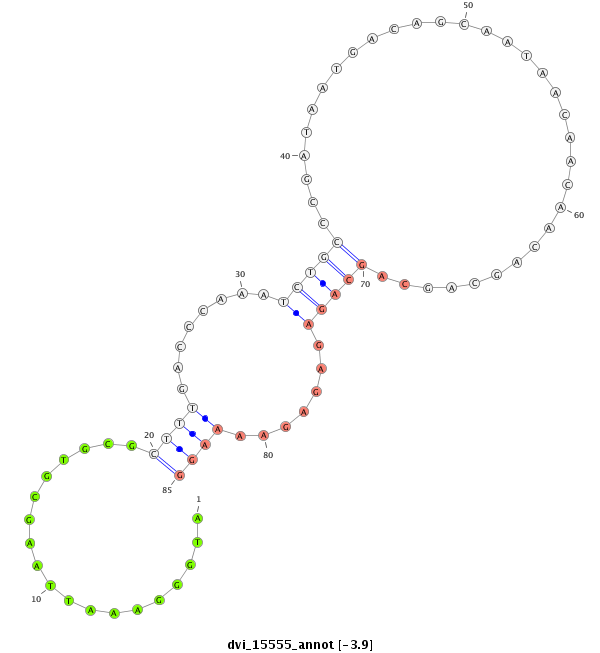
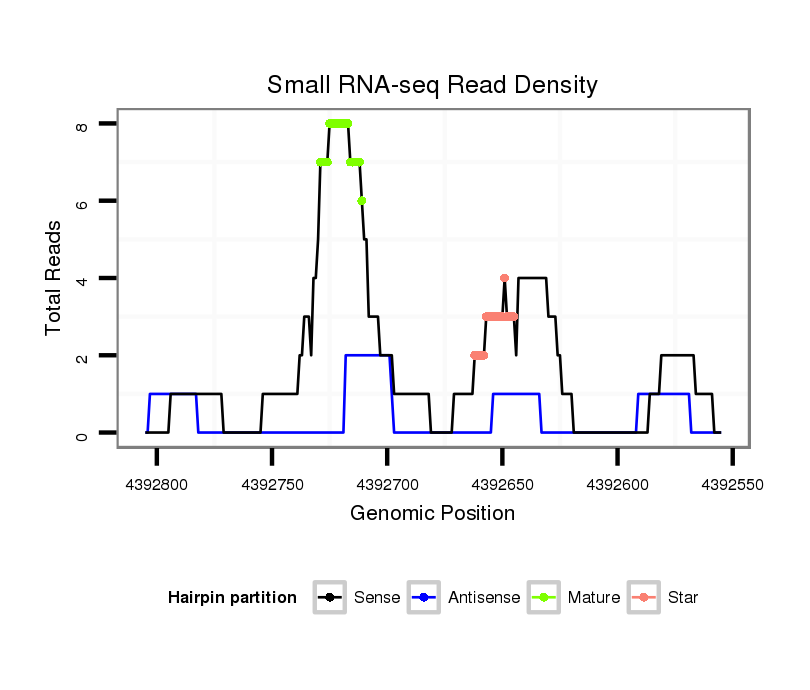
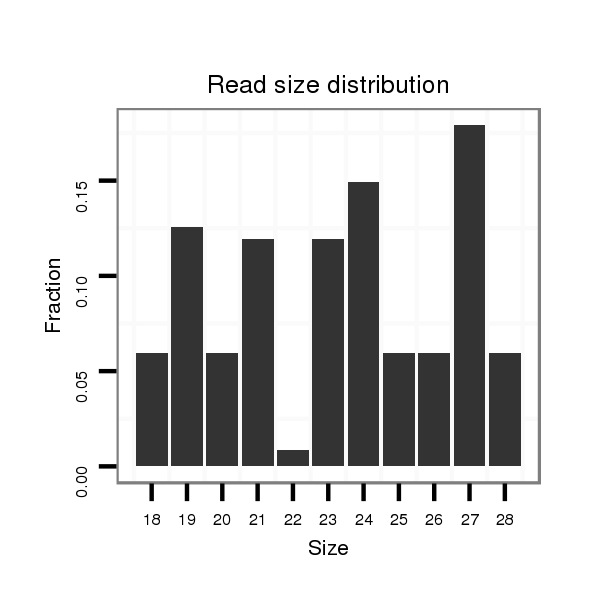
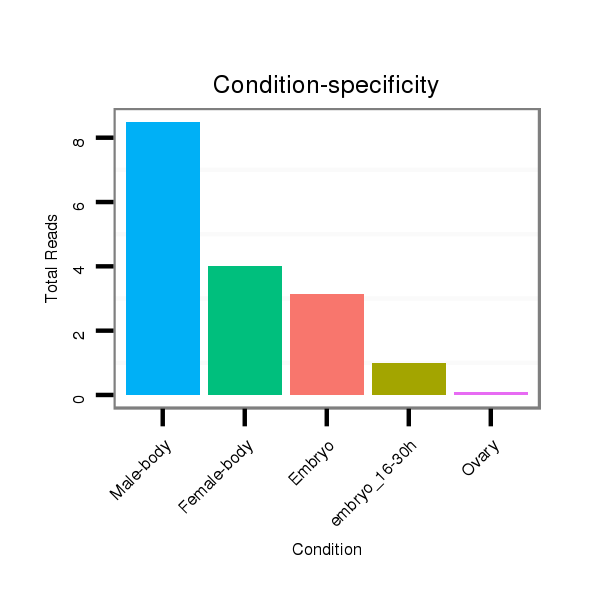
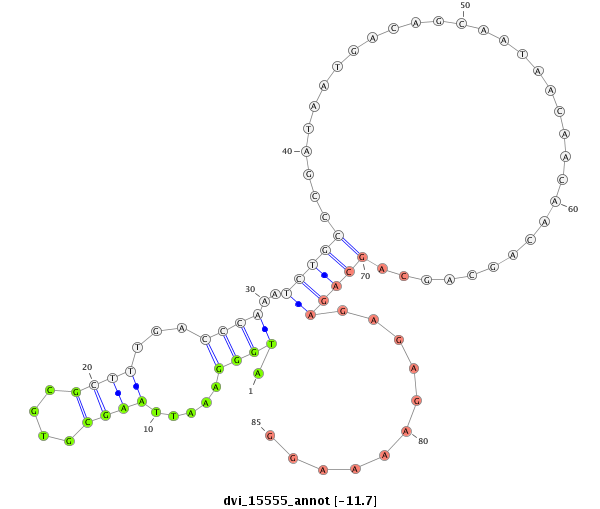
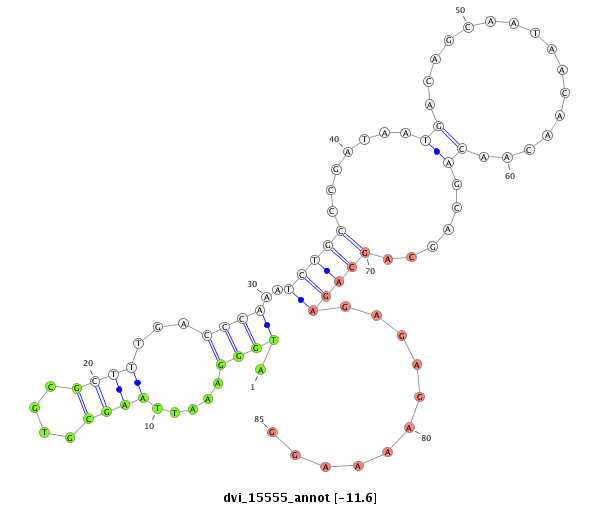
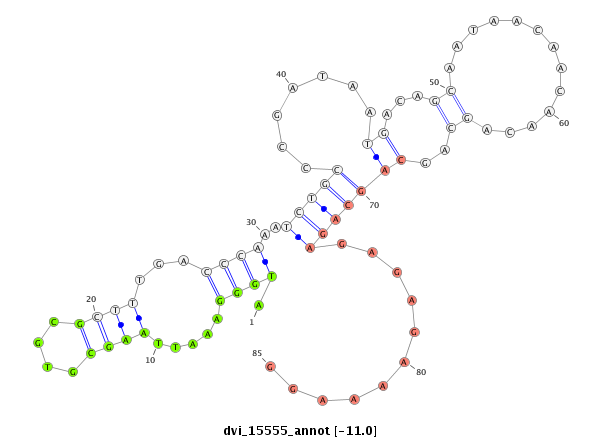
ID:dvi_15555 |
Coordinate:scaffold_13047:4392605-4392755 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
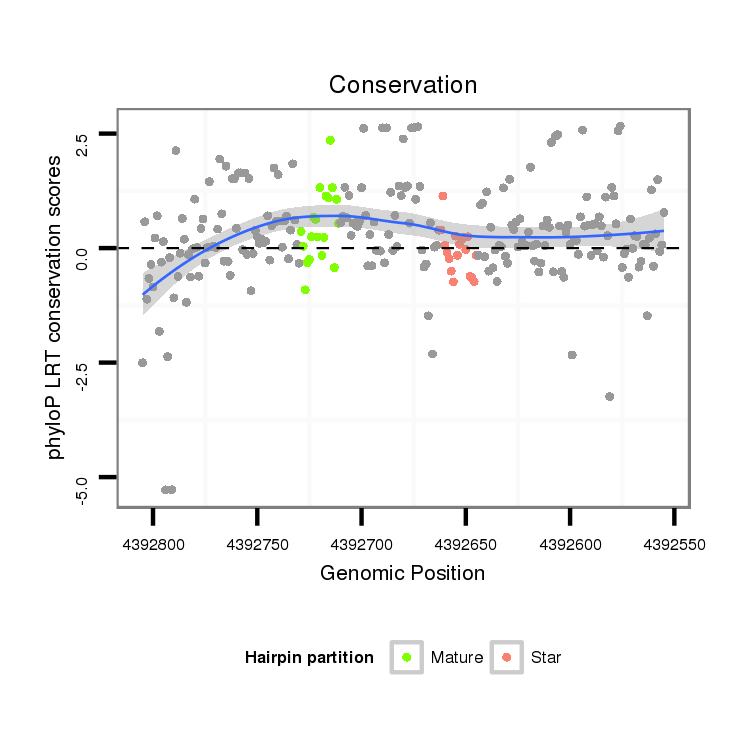
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -11.7 | -11.6 | -11.0 |
 |
 |
 |
CDS [Dvir\GJ23359-cds]; exon [dvir_GLEANR_8716:4]; intron [Dvir\GJ23359-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACAACGGCAGCGACTGCGGCAGGTGTGTCACAGCAGACAGCTTCCAAATTACCACACAACAAAAACAACAGTAATAATGGGAAATTAAGCGTGCGCTTTGACCCAAATCTGCCCGATAATGACAGCAATAACAACAACAGCAGCAGCAGAGAGAGAAAAGGCGAGAACATTGGCGGCAGCTATGACAGCTTGCATGCGAAGGTTGCGCGCAAGGCGCGCTCTCTCAGCAATTCGCCGCTATACAGAAAGAA ****************************************************************************...................((((........(((((.................................))))).......))))****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M047 female body |
M061 embryo |
SRR060665 9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
V047 embryo |
SRR060655 9x160_testes_total |
SRR060664 9_males_carcasses_total |
SRR060687 9_0-2h_embryos_total |
SRR1106730 embryo_16-30h |
SRR060674 9x140_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................ATGGGAAATTAAGCGTGCG............................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TCTCTCAGCAATTCGCCGCT............ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CAGCAGAGAGAGAAAAGG.......................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................ACAGTAATAATGGGAAATTAAGCGTGC............................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................ATGGGAAATTAAGCGTGCGCT.......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAAATTAAGCGTGCGCTTTGACCCAAAT............................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GACCCAAATCTGCCCGATAATGACA............................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GAAGGTTGCGTGCAAGGCG................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................AATGGGAAATTAAGCGTGCGCTTTGAC..................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AAAGGCGAGAACATTGGCGGCAG........................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................CCACACAACAAAAACAACAGT................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GAGAACATTGGCGGCAGCTATGAC................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GACTGCGGCAGGTGTGTCACAGC......................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GAGAACATTGGCGGCAGCT...................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CAACAGCAGCAGCAGAGAGAGAA.............................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AGTAATAATGGGAAATTAAG.................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CAGCAATTCGCCGCTATACAGAA.... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................ATAATGGGAAATTAAGCGTGCGCT.......................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GAAAAGGCGAGAACATTTGCGGC.......................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................ATAATGGGAAATTAAGCGTGCGCTTT........................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GAGAGAGAAAAGGCGAGAACATTGGCG............................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AATAACAACAACAGCAGCAGCAGA..................................................................................................... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................CAACGATGACAGCTTGTATGC...................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................GAACTTTGGCGGCTGGTATG................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................ACAGTCATGATGGGAAATCA.................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AACAACAACAGCAGCAGCAGAG.................................................................................................... | 22 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ACAACAACAGCAGCAGCAG...................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................GAAATTAGGCGAGCGCTT......................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................CTGACCGATGATGCCAGCAA........................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGTTGCCGTCGCTGACGCCGTCCACACAGTGTCGTCTGTCGAAGGTTTAATGGTGTGTTGTTTTTGTTGTCATTATTACCCTTTAATTCGCACGCGAAACTGGGTTTAGACGGGCTATTACTGTCGTTATTGTTGTTGTCGTCGTCGTCTCTCTCTTTTCCGCTCTTGTAACCGCCGTCGATACTGTCGAACGTACGCTTCCAACGCGCGTTCCGCGCGAGAGAGTCGTTAAGCGGCGATATGTCTTTCTT
******************************************************************************************...................((((........(((((.................................))))).......))))**************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060681 Argx9_testes_total |
M061 embryo |
SRR060676 9xArg_ovaries_total |
V047 embryo |
V053 head |
SRR060669 160x9_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
M027 male body |
SRR060664 9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................AAGCGGCGATATGGGTTT... | 18 | 2 | 10 | 17.00 | 170 | 168 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGGCTTT... | 18 | 2 | 3 | 6.33 | 19 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGTGTTT... | 18 | 2 | 20 | 3.00 | 60 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGGGTTTCT. | 20 | 3 | 6 | 1.50 | 9 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGTTTTT... | 18 | 2 | 3 | 1.33 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TCGCACGCGAAACTGGGTTT................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TCGCACGCGAAACTGGGTTTA............................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TTGCCGTCGCTGACGCCGTCC.................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GCGCGAGAGAGTCGTTAAGCGGC.............. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TCTCTTTTCCGCTCTTGTAAC............................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGGGTTTCTT | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................ATTGTCGAAGGTTTAATGG...................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CGAGGGAGTCCTTAACCGGCG............. | 21 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGGCTTTA.. | 19 | 3 | 20 | 0.40 | 8 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| ..................................................................................................................................................................................................................................CGTAAAGTGGCGATATGT....... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AGACTGCCGAACGTACGCTG................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................AGTGGCGTCCGTCGAAGGTGT........................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGTGATATGGCTTT... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................CGGGAGAAAGTCGTTAAG.................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TCGTTAAGCGGGGTAATGTC...... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................AGCGGGGATATGGGTTTCTT | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGCGATATGGGTTC... | 18 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGGGATATGTCTAG... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAGCGGTGATATGGGTTTC.. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................ATTGTTGTTGTCGTCGTCGT....................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:4392555-4392805 - | dvi_15555 | ACA--ACGGCAG-------C---GACT---GCGGCAGGT------------------------GTGTCACA------------GCAGACAGCTTCCAAATTACCACACAACAA-------------------------------------------------------------------------------------AAACAACAGT---------AATAATG------G--------GAAATTAAGCGTGCGCTTTGACC--CAAATCTGCCC---GATAAT---GACAGCAATAACAA---------------------C----------------------A---------------------------------------------------------------------------------ACAGCAGCAGCA------------------------------G--AGAGAGAAA------------AGGCGAGAACATT--GGCG----------------GCA----------------------GCTATGACAGCTTGCATGCGAAGG---TTGCGCGCAAGGCGCGCTCTCTCAGCAATTCGCCGCT-------------------------------ATA---CAGAAAGAA |
| droMoj3 | scaffold_6540:21975884-21976143 + | GCA--ACAGCGG-------C---AGCG---GCAGCGGCT---------------------GGCCTGCCACA------------GCAGACAGCTTCCAAATTACTACACAGCAA-------------------------------------------------------------------------------------CAACAGCAAT---------AACAATG------G--------GAAATTAAGCGTCCGCTTTGACC--CAAATCTGCCC---GATAAT---GACAGCGACAAGAA---------------------C----------------------AACA------------------------------------------------------------------------------ACA---GTAGCA------------------A------AAGCAA--AGAGAGAAC------------CTATGAGAGCTTA--GGCG----------------GCA----------------------GCTATGACAGTTTGCACGCGAAGG---TTGCCCGCAAGGCGCGCTCTCTTAGCAATTCGCCGCT-------------------------------ATA---CCGAAAGAG | |
| droGri2 | scaffold_14906:9876276-9876550 - | ACT--GCAAATG-------C---GAAT---CCGAATCCGAGTGCGAATGCGAATGCGTCAGGTGTGTCACA------------GCAGACAGCTTCCAAATTGCGACAAAGCAG-------------------------------------------------------------------------------------CAACAACAAT---------AATGGAA------A--------CAAATTAAGCGTGCGCTTTGACC--CAAATCTGCCCGATAATAATAATGACAGCAACAAC----------------------------------------------A---------------------------------------------------------------------------------ACAATAACAACA------------A------CAACAACAATTG--CGAGAGCA-----------------------------ACT----------------ACA----------------------ACTATGACAGTTTACATGCGAAGG---TTGCCCGCAAGGCGCGCTCTCTCAGTAATTCGCCGCT-------------------------------GTA---TCGCAAGAA | |
| droWil2 | scf2_1100000004943:15996366-15996607 + | GCA--GCAGCAG-------C---AGCA---GCAACAACA---ACA---------------------ACAAG------------GAATGCAACAACCAAATTGCCCATCAGCAA-------------------------------------------------------------------------------------CAACAAAAATAGTAACAACAACAGCA------A--------CAAATTAAGCGTACGCTTTGACC--CAAATTTACCCGCCGACAAT---GACAACAACAACAA---------------------C----------------------A---------------------------------------------------------------------------------AAAATAGCCACG-------------------------------------------------------------------------ACAATG-----------------------------------GCAACAACAACCAAAATGCAAAAT---CTTCACGCAAAGCTCGCTCTCTGAGCAATTCTCCGCA-------------------------------ATATCATCGAAAGAA | |
| dp5 | 2:25496316-25496579 - | GGC--------------------------------------------------------GGGCGTTCCACATGTTGCTG---------CAACCTCCAAAGTGCTGCAAGGCAGCAGTCTCAGCCCCAGCCCCCACCACCACC------ACCACC----A---------------------------------------------CCAT---------AACGGCC------C--------CAAATTGAGCGTGCGCTTTGACC--CAAATCTCCCCATCGACAAT---GACAGCAACAACAA---------------------C----------------------AGCAATGGAAAG---------------------------------------------------------------------------------A------------G------CATCAGCAGC--------------------------------A---GC--AGCG----------------GCA----------------------GCAGCGAGAGCCAGAATGCAAAAA---TCGGCCGAAAGGCGCGCTCTCTCAGCAATTCTCCGTT-------------------------------GTACCATCGCAAGAA | |
| droPer2 | scaffold_6:824408-824650 - | GGC--------------------------------------------------------GGGCGTTCCACATGTTGCTG---------CAACCTCCAAAGTGCTGCAAGGCAGCAGTCTCAGCCCCAGCCCCC---ACCACC------ACCACC----A---------------------------------------------CCAT---------AACGGCC------C--------CAAATTGAGCGTGCGCTTTGACC--CAAATCTCCCCATCGACAAT---GACAGCAACAACAA---------------------C----------------------AGCAATGGAAAG---------------------------------------------------------------------------------A------------G------CATCAG---------------------------------------------------------------------------------------------CGAGAGCCAGAATGCAAAAA---TCGGCCGAAAGGCGCGCTCTCTCAGCAATTCTCCGTT-------------------------------GTACCATCGCAAGAA | |
| droAna3 | scaffold_12911:2281892-2282061 + | GCA--TCCGCAC-------G---AGCA---T----------------------------------------------------------------------------------------------------------------------CGGGC----G---------------------------------AGAAAA--------------------------CAGCCCC--------CAAATTGGGAGTGCGCTTTGACC--CAAATCTGCCC---GATAAT---GACAGCAACAACAA---------------------T----------------------C---------------------------------------------------------------------------------TGC---CGAGCG-------------------------------------------------------------------------AGAATGCAAA-----------------------------------------------TTCGAAAA---GCGGCCGCAAGGCGCGCTCTCTGAGCAACTCGCCGCT-------------------------------GTACGAGCGCAAGAA | |
| droBip1 | scf7180000396714:798631-798804 - | ACA--TGC--ATCCA----C---GAAG---ACG-------------------------------------------------------------------------------------------------------------------GCGGGC----G---------------------------------AGAAAA--------------------------CAGCCCC--------AAAATTAGGAGTGCGCTTTGACC--CAAATCTGCCC---GATAAT---GACAGCAACAACAA---------------------T----------------------C---------------------------------------------------------------------------------TCC---CGAGCG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TTCGAAAA---GCGGCCGCAAGGCGCGCTCTCTCAGCAACTCGCCGCT-------------------------------GTACGAGCGCAAGAA | |
| droKik1 | scf7180000302690:225569-225759 - | CCG--ACAGCTA-------T---ATCT---GCAGCAACA-----------------------------------------------------------------------------------------------------------------------G---------------------------------CAACAA--------------------------AGCCATC--------CAAATTAAGCGTGCGCTTCGACC--CAAATTTACCCAAAGATAAT---GACAGCAACAACAA---------------------C----------------------C---------------------------------------------------------------------------------TGAACAGCACTG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------TGCGAATTCGAGTTCGAAAG---GCGGACGCAAGGCCCGATCGCTGAGTAACTCGCCGCT-------------------------------GTATGAGCGCAAGAA | |
| droFic1 | scf7180000453812:968186-968452 - | ACA--TAC--GGCAGCG-GCAGCGGCGGCGGCAGCAGCA---GCAACAGCGG--------------------------------TT-GCCACAA-------------CGGTAA-------------------------------------------------------------------------------------CAACGGCAGTGGCAGCGGCAGCGGCA------G--------CGGCGTCGGCGTCGGCAACATTT--GCAATCAGCAGATCAACAAC---TACAGCAATAACAA---------------------C----------------------AACAACGGCAGCAGCAGCAGCAATCATATGAGTGCAGGCGGTTTCTTCGGCGG---------CAGCAGCAGCAGCAGTGGCAAC---------A------------G------CAATACCGACTACTTGGCCACG--------------------------------------------------------------------------------------------------------------------------------CCGACAACC-------------------------------GCTTATGCGACACCA | |
| droEle1 | scf7180000490996:1013959-1014232 + | GCG--ACAGCAG-------C---AACT---GCAGCAGCAGTTGCAGCAGCAG--------------------------------CT-GCAACAG-------------CAATTG-------------------------------------------------------------------------------------CAACAGCAAC-------------------------------------------------------------------------------------------AG---CAACAGCAGCAGCAGCAACCATCGCCCATGTTGGGCAGACCAGCGGCAGCCACGCCC---------ACAGATATGCA-GCGTTATGTGCAGAGGATGCAACAACAACAGCAGCAACAGCAGCAACA---GCAGCA------------------A------CAGCAG--CAG-----------------------------------CA----------------GCA----------------------GCAACAGGAGCTGTACCGCC----------------------------------AGCAACAGCAGCAACATCGCACTAGTATGAGCGGCATAGCCAGC---CAGCAACAC | |
| droRho1 | scf7180000760202:71820-72072 + | ACA--ACAGCAG-------C---AACA---ACAGCAGCA---GCA---------------------ACAAC--------AACAGCA-GCAGCAA-------------CAA------------------------------------------------------------------------------------------------------------------CAGCAGCA-----------------------------------------GCAAC---------------------AA---CAGCAGCAGCAACAACAAC----------------------AGCAGCAGCAACAACA---------------------------------GCAGCAGCAACAACAACAGCAGCAGCAACAACAACAGCAGCAGCAACAACAGCAGCAGCAACAACAGCAG--------CAA-----------------------------------CA----------------GCAACAACAACAACAGCAGCAACAG-CAAC------------------------AACAACAACAGCAGCTGCAACAACAACATCAGGTC-------------------------------TTGGCACAGCAACAA | |
| droBia1 | scf7180000302155:399668-399858 + | CTCCACCA--T------G-T---GG---------------------------------------------------CCG---------C-----------------------------------------CCCACCTGCG--------CCCGCC----C---------------------------------CCAAAA--------------------------CACCACC--------CAAACTGAGCGTACACTTTGACC--CCAATCTGCCCAAAGATAGCAGCGACAGCAACAACAA---------------------C----------------------T---------------------------------------------------------------------------------TAA---GCAGTG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TGCCAAGAGCGGCGGCCGCAAGGCGCGTTCGCTGAGCAATTCGCCGCT-------------------------------GTACCACCGCAAGAA | |
| droTak1 | scf7180000415417:340804-340986 - | CCA--ACA--T------G-C---CG---------------------------------------------------CCG---------C-------------------------------------------------CACTGTCCCCGCCGCC----C---------------------------------CTAAAA--------------------------CACCACC--------CAAATTGAGCGTACACTTTGACC--CCAATTTGCCCAAAGACAAC---GACAGCAACAACAA---------------------T----------------------A---------------------------------------------------------------------------------TTA---GCAGCG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TGCTAAAA---GCGGTCGCAAGGCGCGCTCGCTGAGCAATTCGCCGCT-------------------------------GTATCATCGCAAAAA | |
| droEug1 | scf7180000409542:105515-105690 - | CCCCAATA--T------G-C---CACC---GCC-------------------------------------------------------------------------------------------------------------------GCCGCT----C---------------------------------CGAAAA--------------------------CGCCGCC--------GAAATTGAGCGTACACTTTGACC--CCAATTTGCCCAAAGACAAC---GATAGCAACAACAA---------------------T----------------------C---------------------------------------------------------------------------------TTA---GCAGTG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TGCCAAAA---TCGGTCGCAAGGCGCGCTCGCTCAGCAATTCGCCGCT-------------------------------GTATCATCGCAAAAA | |
| dm3 | chr3R:17030900-17031093 + | CCTCAATA--T------G-C---CAAC---CC-------------------------------------------------------------------------------------------------TCCCGCCGACACT------GCCGCC----C---------------------------------CGAAAA--------------------------CGCCACC--------CAAGCTGAGCGTACACTTTGACC--CCAATTTGCCGAAAGACAAC---GATAGCAACAACAA---------------------C----------------------C---------------------------------------------------------------------------------TCACCGGCAGCG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TGCTAAAGGCGGCGGTCGCAAGGCGCGTTCACTAAGCAATTCGCCACT-------------------------------GTATCACCGAAAAAA | |
| droSim2 | x:11775184-11775462 - | ACA--TCCGCAG-------C---AGCA---GCGGCAAAC---GCA-----------------------ACA------------GCA-GCAGCTCCCCAACTACCGC-----------------------CCCC---------------GCTGCCCGTTACCGTGGCAACATTGCCCACTCAGGCGGCGACGG------CAGCAGCAGC---------AACAGCT------G--------CG------------ATTTTGGCC------ACTCACCAGCAACAGC---AGCAGCAGCAACA-------------------------------------------------------------------------------------------------------------------------------ACAGCAGCAGCAGCAATTGCAGCAG------CATCAGC--------------------------------------ATTTGA--------GCA--GCAGCAACAACAACAACAACGGCGGCAGCAC-ACAC------------------------GGCCAAC---AGCATACGCACAGCAACA--ACAAC-------------------------------AACACATTGCCAGTA | |
| droSec2 | scaffold_0:13819818-13820029 - | ACA--ACGGCAG-------C---GGCG---GCAGCATTT---GCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCAGCAGATCAACAAC---TACGGCAACAACAACAACGTCGGC-------------------------------------------------------AATCATATGAGTGCAGGCAGTTTCTTCGGCGGGGCCAACAGCAACATCCACAGTAGTGGCAAT---------A------------G------CAATACCGATTATATGACCACGCCAACCACCGCTTATGCGACA---------------------------------CCAGCGACGGCAGCCACAT-CCAC------------------------GGTGAAC---A-----------------------------------------------------------------CCACA | |
| droYak3 | 3R:5815705-5815896 + | ACA--ATA--T------GGCAATCC--------------------------------------------------------------------------------------------------------CCCCGCCGACACT------GCCGCC----A---------------------------------CGAAAA--------------------------CGCCGCC--------CAAGTTGAGCGTACACTTTGACC--CCAATTTGCCAAAAGACAAC---GATAGCAACAACAA---------------------T----------------------C---------------------------------------------------------------------------------TCGCCGGCAGCG-------------------------------------------------------------------------AAAATGCAAA-----------------------------------------------TGCTAAATGCGTCGGCCGTAAGGCGCGTTCGCTGAGCAATTCGCCACT-------------------------------GTATCACCGCAAAAA | |
| droEre2 | scaffold_4690:4615009-4615231 + | ACA--ACGACAA-------C---GAAA---ACAACAACA---GCA---------------------AGCA-------------------------------------CAGCAG-------------------------------------------------------------------------------------CAGCAGCAAC---------AA-----CAACAACAAGTGTAGTGGGTCGCACGCGCGACTTGTGTGTGTGAATAATGAAGAAAAAGC---AGAAGCAACAACAA---------------------C----------------------AGCAGCAGCAGCAGCA------------------------------------------------------------------GCA---GCAGAG-------------------------------------------------------------------------GCAGAGCAAGGCGCCATCTG------------------------------------------AAG---CGGAATCGGAGGCAGACCGCCCCGGC-AGCAACAGCA-------------------------------GCAGCACCAACAACA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 07:16 PM