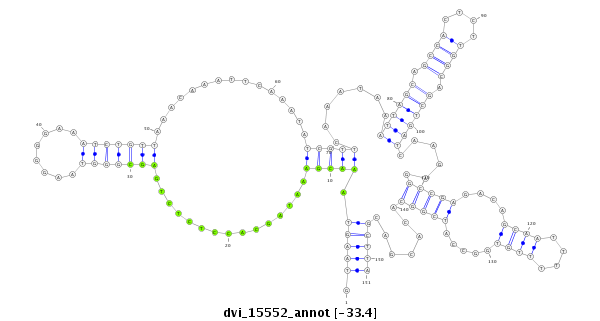

ID:dvi_15552 |
Coordinate:scaffold_13047:4390429-4390579 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
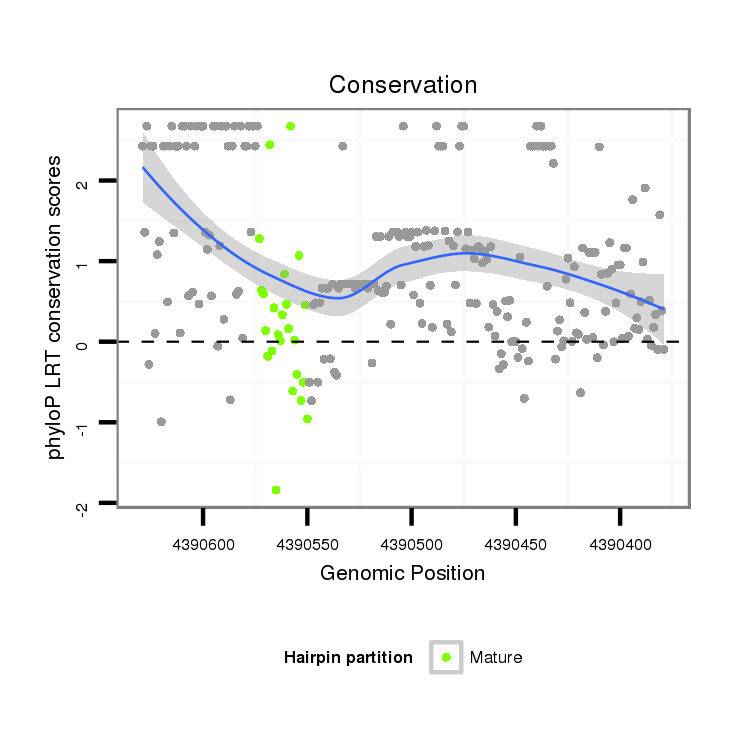
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.8 | -32.8 | -32.8 |
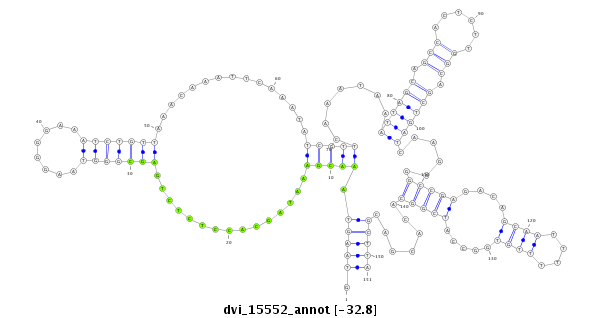 |
 |
 |
intron [Dvir\GJ23359-in]; CDS [Dvir\GJ23359-cds]; exon [dvir_GLEANR_8716:5]; intron [Dvir\GJ23359-in]
No Repeatable elements found
| -#################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTTACCCCGGGCCGACCCCTTCTTGGACAAAGTCAATCTATCAGATTTCGGTAAGTAAACGAAATAGCACCTCTCTGAGCGGGTAAGGGGAAATCTGTTAAACAAATTCAAATATCGTTCAAATAAATTAGCAGCCACTCTTGGCAGCTGATCAAGAGGCCGAGACAGCAATTTTTTGTGGCCATCGGCACACGACGCTTAGCCAACCACTAAGAACAGACATGTATTATGTACATATATATTTTTTGATT **************************************************.(((((.(((((...............(((((((........)))))))...............))))).......((((((.((((....)))).))))))......(((((....((((....)))).....))))).......)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1106730 embryo_16-30h |
M047 female body |
M027 male body |
SRR060665 9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................AAACGAAATAGCACCTCTCTGAGC........................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TACCCCGGGCCGACCCCTTCTTG.................................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................TTGGACAAAGCCAATCTATCAGAT............................................................................................................................................................................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CCATCGGCACACGACGCTTAG................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................AGTCAATCTATCAGATTTCGAGGA..................................................................................................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........GGCCGACCCCTTCTTGGAC............................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...ACCCCGGGCCGACCCCTTCTTGGA................................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CTTGGCAACTGACGAAGAGGC........................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................AGAGAGCGATTTTTTCTGGC..................................................................... | 20 | 3 | 7 | 0.43 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| ............................................................................GAGCGGGTAACGGGGAAT............................................................................................................................................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CAATGGGGCCCGGCTGGGGAAGAACCTGTTTCAGTTAGATAGTCTAAAGCCATTCATTTGCTTTATCGTGGAGAGACTCGCCCATTCCCCTTTAGACAATTTGTTTAAGTTTATAGCAAGTTTATTTAATCGTCGGTGAGAACCGTCGACTAGTTCTCCGGCTCTGTCGTTAAAAAACACCGGTAGCCGTGTGCTGCGAATCGGTTGGTGATTCTTGTCTGTACATAATACATGTATATATAAAAAACTAA
**************************************************.(((((.(((((...............(((((((........)))))))...............))))).......((((((.((((....)))).))))))......(((((....((((....)))).....))))).......)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M027 male body |
SRR060684 140x9_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060657 140_testes_total |
SRR060662 9x160_0-2h_embryos_total |
M047 female body |
M028 head |
SRR060687 9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TGGTGATTCTTGCCGGTA............................ | 18 | 2 | 1 | 7.00 | 7 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TGGTGATTCTTGCCGGTAC........................... | 19 | 2 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TGGGCTGGGAATCGGTTAG........................................... | 19 | 3 | 14 | 1.57 | 22 | 1 | 0 | 0 | 13 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTGATTCTTGCCGGTAC........................... | 18 | 2 | 7 | 1.57 | 11 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GGAAGAACCTGTTTCAGTTAG..................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TGGTGATTCTTGCCGGTACG.......................... | 20 | 3 | 9 | 1.00 | 9 | 6 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGTGGTGATTCTTGCCGGTA............................ | 20 | 3 | 5 | 0.80 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGTGGTGATTCTTGCCGGTAC........................... | 21 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGAACGGTCGACTAGATC............................................................................................... | 18 | 2 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TGGAGAAGACCCTGCTTCA.......................................................................................................................................................................................................................... | 19 | 3 | 7 | 0.29 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGAACGGTCGACTAGATCT.............................................................................................. | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCCGGCTCTGTCGTTACG............................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GTTGATTCTTGTCGGTAC........................... | 18 | 2 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CTGGTGAAGAACTTGTTTG........................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................TGGTTGACTTTATAGCAAGT.................................................................................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TTTTTGATTCTTGTCTGTACG.......................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AAAAAACACCCGTAGCCGTCTT.......................................................... | 22 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TTCTTGGGAAGAACCTGTT............................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................TAGGAAGTTTATTTTAGCGT...................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:4390379-4390629 - | dvi_15552 | GTTACCCCGGGCCGACCCCTTCTTGGACAAAGTCAATCTATCAGATTTCGGTAAGTA------AACGAAATAGCA----------------------------------------------------------------------------------------------------------CCTCTCTGAGCGGGTAAGGGGAAATCTGTTAAAC-AAATTCAAATATCGTTCAAATAAATTAGCAG-CCACTC----TTGGCAGCTGATCAAGAGGCCGAGACAGCAAT-------T----------T-------TTTGTGGCCA-----TCGGCACACGACG---CTT-AGCCAACCACTAAGAACA--------GAC------AT--------------------------------------------GTATTATGTACATAT----A--TATTT-------------------TTTG----ATT |
| droMoj3 | scaffold_6540:21978104-21978408 + | GTTGCCGCGTGCCGATCCCTTCCTGGACAAAGTAAACCTTTCAGATTTCGGTAAGTA------AACCAAAAA---------------------AGAAACCGG---------------------------------------------------------------------------GG--GCTCGTTTGGGATGTGAGAGGAAAACTGTTAAAC-AAATTCATATATCGTTCAAATAAATTAGCAG-CTACAC----TTGGCAACTGGCCAAGAAGAGGC---TGAGGCATCAATATTTCTTTCTTTCTT---TTTA----TCTG-----TCGGCACACGTCG---CGACACTTAACCACGAAGAACAGATGTATAGA--------------TAT--------------TA-TATAAATAAATAAAT-AT-GTA-TATGTACATATATGTGGGTA-TT-------------------TTTG----ATT | |
| droGri2 | scaffold_14906:9873868-9874168 - | GTTACCCCGTGCCGATCCTTTCTTGGACAAAGTCAATCTATCAGATTTCGGTAAGTA------AACGAAACATTCCCA--AAACGATCC--CCCTTG-CCA-----------------------------AAGAG-----------------------------------------TGG--GATGGTT----------GGGGGACACTGTTAAACTAAATTCAAATATCGTTCAAATAAATTAGCAG-TCACAC----TTGGCAACCGATCAAGT---GGCCAACGAGAT---AATTCTTTTTTTTTTCTTTTTTTTTT---GTGT-----TGGGCACACGTCG---CTTTAGC-A-CATT--TGAATA--------GAAACGTACCGAAACCTA---------------TA-TTTAAATAAAT---------------------------A--T---------ATATGTATAC---GCATA----ATT | |
| droWil2 | scf2_1100000004943:15998978-15999232 + | GTTGCCCGTTGCCGATCCGTTCCTAGAGAAAGTCAATCTATCAGATTTTGGTGAGTA------TTAG---------------------------------------------------------------------------------------CTAGAAATTTGATGGGAATTAAGGG--GCTGGCT--------------------G----------------GTGGACGTGGGGGAGGGGATGA-GCACAA----TTGGCAGTTGATCAAGAGTCTACCTCTGCGAG-------A------------------T-----GCTAAAAACAAGGCACACGACG---CTGTCGTCA-AAGC--T-------------AAA------TATT-CCTCATTTATACAAATACATAT-------------AT----GTA----------------A--TATATACCAATATATACGCA---ATTTG----ATT | |
| dp5 | 2:25494298-25494506 - | GCTGCCTGTAGCCGATCCGTTCCTAGAGAAGTGCAATCTATCTGATTTTGGTAAGTATTATTCTTCGATCGCACCTCG--CAACCTGC---CACATGCCCCGTGCC------------------CCGCCTCTGAGGCGCA-CCAT-GGGGCAAAC----------------------AAAAAAT------------------------G----------------TTTAATTTAAATAAATTAGCAG-ACACAC----TTGGCATTCGATCAAGAGTCGGCCTCCA--------------------------------------TT-----GTGGCACACGACG------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_6:822398-822606 - | GCTGCCTGTAGCCGATCCGTTCCTAGAGAAGTGCAATCTATCTGATTTTGGTAAGTATTATTCTTCGATCGCACCTCG--CCACCTGC---CCCATGGCCCGTGCC------------------CCGCCTCTGTGGCGCA-CCAT-GGGGCAAAC----------------------AAAAAAT------------------------G----------------TTTAATTTAAATAAATTAGCAG-ACACAC----TTGGCATTCGATCAAGAGTCGGCCTCCA--------------------------------------TT-----GTGGCACACGACG------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_12911:2283838-2284120 + | GTTGCCTGTCGCCGATCCGTTCCTAGAGAAAGTAAACCTTTCGGACTTGGGTAAGTT------TATGAGCAGCCCCCG--TATTCCCAGCCCCCCCAACCCCCCT----------------T--CAGTGCCA--TTGGCC-GATG-GGCACAAAC----------------------AA-AAAT------------------------G----------------TTTAAATTAAATAAATTAGCAG-ACACAC----TTGGCATTCGATCAAGAATCGGCCTCTGATGG-------T------------------GC----TCTG-----TGGGCACACGACGACGCCATCCCCT-GAGC----------------TAA------TAGGACCTACTATAT--------ATCT-------------GTGGGG--------------T----A--TAT------AAGAGCAATACTAGAATTCATG---- | |
| droBip1 | scf7180000396714:796524-796742 - | GTTGCCTGTCGCCGATCCGTTCCTAGAGAAAGTAAATCTTTCGGACTTGGGTAAGTT------TTTGAGCAGCCCCCG--TGTCCCCAA--CCCCCAACCCCCAACCCCCAACCCGCCCCTC--CAT--------CCCC--ACCG-GGCACAAAC----------------------AA-AAAT------------------------G----------------TTTAAATTAAATAAATTAGCAG-ACACAC----TTGGCATTCGATCAAGAATCGGCCTCTGATGG-------C------------------TT----TCGG-----TGGGCACACGACG------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302690:222015-222269 - | GTTACCTGTCGCTGATCCGTTCCTAGAGAAAGTAAATCTATCTGATTTGGGTAAGTT------TTCAAACCACCCTCT--CAAATGGCG--TGCATTTCCACC---------------------GTACC--C--CTTCCC-CCTA-AGAATATAG----------------------AA-AAAT------------------------G----------------TTTAATTTAAATAAATTAGCAA-ACACAC----TTGGCATTCGATCAAGAGTCGGCCTCTGATGC-------C------------------GT----TCTG-----TGGGCACACGACG---CTATCGCCA-GAACTAATAACA--------GA--------AAAACCTGCCAG------------------------------GGG-T-----ATACATAT----A--TA--------------------------------- | |
| droFic1 | scf7180000454096:310528-310808 + | GTTGCCTGTTGCTGATCCGTTCCTTGAGAAAGTAAACCTATCTGACCTGGGTAAGTT------TTCGATTAACCCTCG--TATTAAGCA--CCTATGTCCCTCACT----------T----CCTTCACC--T--TGCCCA-CCTTGGGAACAAAC----------------------AA--AAT------------------------G----------------TTTAATTAAAATAAATTAGCAAAACACTC----TTGGCATTCGATCAAGAGTCGGCCTCTGAGGA-------T------------------T-----TCTT-----CGGGCACACGACG---CCATCGCCA-CAAA-AA-------------AAA------CAAAATCTAC-AT--------------------------------------ATGTATATAC----A--AATCGGTTGAAGAAGCAGAC---GAGCTATG---- | |
| droEle1 | scf7180000491080:907276-907560 + | GTTGCCTGTTGCTGATCCCTTCCTAGAAAAAGTAAATCTTTCGGACCTGGGTAAGTT------TTCAAACCGCCCTCT--CATTCTACA--CTCTTATCCCCTGTTGCG---------------CCACC--T--GTCTCCTCTTT-GAGACAAAC----------------------AA-AATT------------------------G----------------TTTAATTTAAATAAATTAGCAA-ACACAT----TTGGCATTCGATCAAGAGTCGGCCTCTGAGGA-------T------------------TT----TCTT-----TGGGCACACGACG---CCATCGGCA-GAAT--A-------------GAA------TAGAACCTAGTATATACGT----ATAT------------------------------ATAT----A--TATAGATCGAAAAAGAAGAC---AAGTTATG---- | |
| droRho1 | scf7180000779250:191645-191886 - | GTTACCGGTTGCCGATCCGTTCCTAGAAAAAGTAAATTTATCGGACTTTGGTAAGTT------TTCAAACAGCCCTCT--CATTCTGCA--CCCATGTCCCGTCCT------------------CTACC--T--GCCTCCTCTTC-GGGACAAAC----------------------AG-AAAT------------------------G----------------TTTAATTAAAATAAATTAACAA-ACACAC----TTGGCATTCAATCAACAGTCGGCCTCTGAGGA-------T------------------TT----TCTT-----TAGGCACACGACG---CCATCAGCA-GAAT--A-------------GAA------TAAAACCTAC-AT----------------------------------------------AT----A--T---------------------------------- | |
| droBia1 | scf7180000302155:402092-402369 + | GTTGCCTGTCGCTGATCCGTTCCTAGAGAAAGTAAACCTATCGGACCTGGGTAAGTT------TTCGACCTACTCTCTCCTGTTCCGCC--CCCTC-TCCCTC-CT---------------A--CCACAACT--GCCGCC-CCTT-GAGGCAAAC----------------------AA-AAAT------------------------G----------------TTTAATTTAAATAAATTAGCAA-ACTCAT----TTGGCATTCGATCAAGAGTCGGCCTCTGAGGA-------T------------------TT----TCTT-----TGGGCACACGACG---CCATAGCCA-GAAT--A-------------GAA------TAGAACCTGGTATAT--------ATGC------------------------------ATGC----T--GATAGACCAAAGAGGAAGAC---ATG------ATA | |
| droTak1 | scf7180000415417:338472-338741 - | GTTACCTGTGGCTGATCCGTTCCTAGAGAAAGTAAACCTATCGGACCTGGGTAAGTT------TTCGAACCACCCTCT--TCTTCCGCT--CCCATGTCCC------------------------------T--GCCGCT-CCGT-GAGACAAAC----------------------AA-AAAT------------------------G----------------TTTAATTTAAATAAATTAGCAA-ACACAC----TTGGCATTCGATCAAGAGTCGGCCTCTGAGGA-------T------------------TT----GCTT-----TGGGCACACGACG---CCATCGCCA-GAAT--A-------------GAA------TAGATCCTGATATAT--------ATAC---------AT-------------------ATAT----A--GGTAGGCCGAAGAAGAAGAC---AAGGTATG---G | |
| droEug1 | scf7180000409542:103069-103330 - | GTTGCCTGTCGCTGATCCGTTCCTAGAGAAAGTAAATCTATCTGACCTGGGTAAGTT------CTCAAACCACCCTCT--CAACGCACC--CCTACT---------------------------TCACC--T--GCCGCC-CCTC-GAGATAAAC----------------------AA-AAAT------------------------G----------------TTTAATTTAAATAAATTAGCAA-ACACAC----TTGGCATTCGATCAAGAGTCGGCCTCTGAGGA-------T------------------TT----GCTA-----TGGGCACACGACG---CTA-------GAAT--A-------------GAA------TAAAACATCCTATAT--------ATAC------------------------------CTAT----A--TATAGGTCCAAGAGGCAGAC---AAGTTATG---- | |
| droSim2 | 3r:4277188-4277480 - | dsi_15260 | GTTGCCTGTCGCTGATCCGTTCCTAGAGAAAGTAAATCTATCAGACCTGGGTAAGTT------TTGAAACCACCCTCT--TAATCCACT--CCCATGTCCCCTTTT------------------CCGCAACG--GCCGTC-CCGA-GAAACAAAC----------------------AA-CAAT------------------------G----------------TCTAGTTTAAATGAATTAGCAA-ACATACTTGGCTGGCGTTTGGTCAAGAGTCGGCCTCTGAGGA-------T------------------TT----TCGT-----TGGGCACACGACG---CCATCGCCA-GAAT--A-------------TAA------TAGAACCTGATAT--GCCAATATATCT------------------------------ATGT----G--TATAGACCGAAGAAGAAGAT---AAGTTGTGGAAT |
| droSec2 | scaffold_38:99314-99527 - | GTTGCCTGTCGCTGATCCGTTCCTAGAGAAAGTAAATCTATCAGACCTGGGTAAGTT------TTGAAACCACCCTCT--TAATCCACT--CCCATGTCCTCT-T----------------T--CCGCAACG--TCCGTC-CCGA-GAGACAAAC----------------------AA-CAAT------------------------G----------------TCTAGTTTAAATGAATTAGCAA-ACATACTTGGCTGGCGTTCGGTCAAGAGTCGGCCTCTGAGGA-------T------------------TT----TCGT-----TGGGCACACGACG---C--------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/19/2015 at 07:15 PM