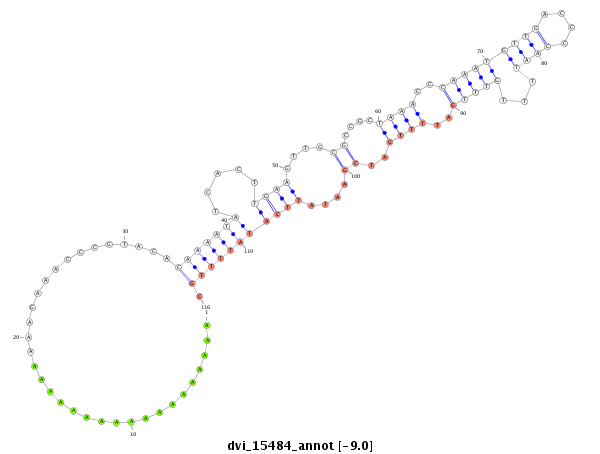




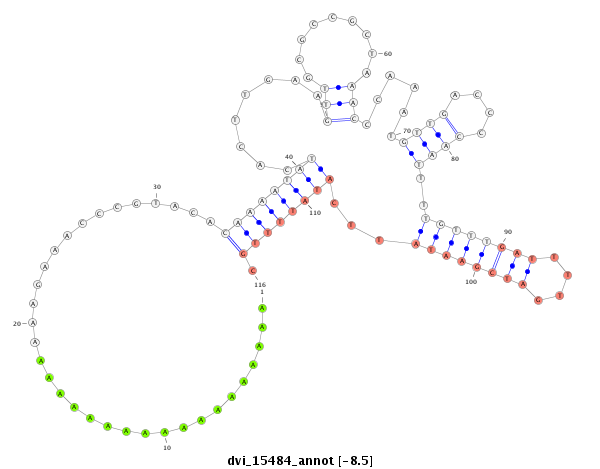
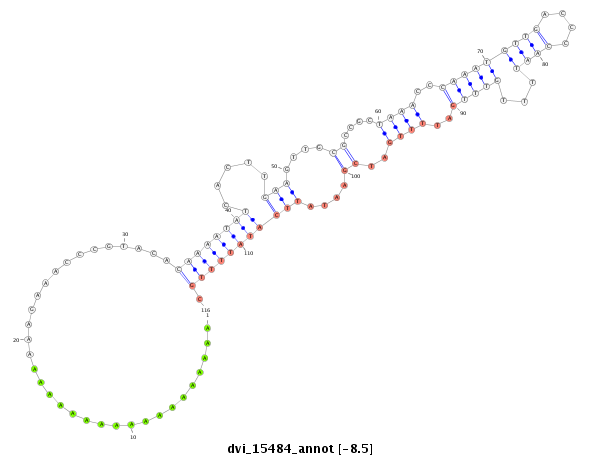
ID:dvi_15484 |
Coordinate:scaffold_13047:4027076-4027226 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.5 | -8.5 | -8.4 |
 |
 |
 |
exon [dvir_GLEANR_9160:5]; CDS [Dvir\GJ23835-cds]; intron [Dvir\GJ23835-in]
| Name | Class | Family | Strand |
| (A)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAAACTGACGACGCGTGGAGGTGAACTGAGCTGAAGTACCATAATAAATGGAATAAATGCGTGAAAATGTGTGCACACAAATAAAAAAAAAAAAAAAAAAAAAAGAAACCCGTACACAAAATATCACTTGAAGTTGCGCCGCTAAACCCAAATGTTGACCCCAATTTTGTTTGATTTTGATCGAATATTCATATTTTGCAGAGCCGGCACGAGAATGGTACGGGCGGCATTGGCAAGAGCGCCAGTGATGT ***********************************************************************************.................................(((((((.....((((....((....((((..(((((((((....))))...)))))..))))..))....))))))))))).**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060661 160x9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
M061 embryo |
V116 male body |
M047 female body |
SRR060669 160x9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060683 160_testes_total |
SRR060689 160x9_testes_total |
V047 embryo |
SRR1106729 mixed whole adult body |
SRR060673 9_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060655 9x160_testes_total |
V053 head |
SRR060666 160_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................AAAAAAAAAAAAAAAAAA...................................................................................................................................................... | 18 | 0 | 20 | 2.40 | 48 | 48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................GATTTTGATCGAATATTCATATTTTGC.................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GAGAATGGTACGGGCGGCATTGGCAAG.............. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................ATGGTACGGGCGGCATTGGC................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................CACGAGAATGGTACGGGCGGCATTGGC................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................ATTGGCTAGAGCGCCGGT..... | 18 | 2 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................AATTGGCTAGAGCGCCGGT..... | 19 | 3 | 18 | 0.33 | 6 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................AGAATGGTACGGGTGGCACTT................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GCACACAAATAATAAAAAAAAAA............................................................................................................................................................ | 23 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GGCGGAATTGGCAGGCGCGC......... | 20 | 3 | 14 | 0.21 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................ATTGGGAAGAGCACCGGTG.... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TAAAAAAAAAAAAAAAAAAAAAAGA................................................................................................................................................. | 25 | 0 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................AAAAAAAAAAAAAAAAAA..................................................................................................................................................... | 18 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CCCGTAGACAAAATTTGACT........................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................ATTGGCTAGAGCGCC........ | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................ACGCAAATGTTGACCGCA........................................................................................ | 18 | 2 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GGAATTGGCAGGCGCGCCA....... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AAATAAAAAAAAAAAAAAAAAAAAA.................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTAATAAAAAAAAAAAAAAAAAAAAAA................................................................................................................................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................AAAAAAAAAAAAGAAACC............................................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AATAAAAAAAAAAAAAAAAAAAAAA................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AATAAAAAAAAAAAAAAAAAAAAA.................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................ATAAAAAAAAAAAAAAAAAA....................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................ATAAAAAAAAAAAAAAAAAAA...................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................TAAAAAAAAAAAAAAAAAAAA..................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTTGACTGCTGCGCACCTCCACTTGACTCGACTTCATGGTATTATTTACCTTATTTACGCACTTTTACACACGTGTGTTTATTTTTTTTTTTTTTTTTTTTTTCTTTGGGCATGTGTTTTATAGTGAACTTCAACGCGGCGATTTGGGTTTACAACTGGGGTTAAAACAAACTAAAACTAGCTTATAAGTATAAAACGTCTCGGCCGTGCTCTTACCATGCCCGCCGTAACCGTTCTCGCGGTCACTACA
****************************************************.................................(((((((.....((((....((....((((..(((((((((....))))...)))))..))))..))....))))))))))).*********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V053 head |
V116 male body |
SRR060667 160_females_carcasses_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060665 9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR060658 140_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
V047 embryo |
M027 male body |
M061 embryo |
SRR060682 9x140_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................AAAGCGGCGATATGGGTTT................................................................................................... | 19 | 2 | 2 | 25.00 | 50 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATTTGGGTTT................................................................................................... | 19 | 2 | 1 | 18.00 | 18 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAGCGGCGATATGGGTTT................................................................................................... | 18 | 2 | 10 | 17.00 | 170 | 168 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAGCGGGGATTTGGGTTT................................................................................................... | 18 | 2 | 4 | 10.00 | 40 | 38 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AACGCGGGGATATGGGTTT................................................................................................... | 19 | 2 | 2 | 6.50 | 13 | 3 | 2 | 1 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAGCGGGGATTTGGGTTTA.................................................................................................. | 19 | 2 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AACGCGGGGATATGGGTTTA.................................................................................................. | 20 | 2 | 2 | 3.00 | 6 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATTTGGGTTTA.................................................................................................. | 20 | 2 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAGCGGCGATATGGGTTTA.................................................................................................. | 19 | 2 | 2 | 2.50 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGCGATATGGGTTTA.................................................................................................. | 20 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATATGGGTTTAC................................................................................................. | 21 | 3 | 18 | 1.06 | 19 | 18 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GGGGTTAAAACAAACTAAAA......................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AACGCGGTGATATGGGTTT................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................CCTCCACTTGACTCGACTTCG...................................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGCGGGGATTTGGGTTTA.................................................................................................. | 18 | 2 | 5 | 1.00 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATTTGGGTT.................................................................................................... | 18 | 2 | 19 | 0.89 | 17 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGCGATATGGGTT.................................................................................................... | 18 | 2 | 17 | 0.82 | 14 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AACGCGGGGATATGGGTT.................................................................................................... | 18 | 2 | 18 | 0.72 | 13 | 7 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GAAAGCGGCGATATGGGTTT................................................................................................... | 20 | 3 | 20 | 0.50 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATTTGGGTTTAA................................................................................................. | 21 | 3 | 6 | 0.50 | 3 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AAGCGGGGATATGGGTTTACAA............................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................ACGCGGGGATCTGGGTTT................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCAACGCGGGGATACGGGTTT................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGTGATATGGGTTTA.................................................................................................. | 20 | 3 | 20 | 0.45 | 9 | 5 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGCGGCGATATGGGTTTA.................................................................................................. | 18 | 2 | 7 | 0.43 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAAAGCGGGGATATGGGTTTA.................................................................................................. | 21 | 3 | 16 | 0.31 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GAACGCGGGGATATGGGTTT................................................................................................... | 20 | 3 | 19 | 0.26 | 5 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................TGATTCGACTTAATGGTA................................................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATGTGGGTTTA.................................................................................................. | 20 | 3 | 19 | 0.21 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGGGATCTGGGTTTA.................................................................................................. | 20 | 3 | 19 | 0.21 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................TTTTTTTCTCTAGGGGCATG........................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GCGGCGATATGGGTTTAA................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AATGCGGGGATATGGGTTTA.................................................................................................. | 20 | 3 | 20 | 0.15 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GAAAGCGGGGATTTGGGTTT................................................................................................... | 20 | 3 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GAAAGCGGGGATTTGGGTTTA.................................................................................................. | 21 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCGCGGGGATGTGGGTTT................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GAACGCGGGGATATGGGTTTA.................................................................................................. | 21 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CACCACGGCGATTTGGATT.................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAGGCGGGGATATGGGTTTA.................................................................................................. | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCAAAGCGGGGATATGGGTT.................................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GGGCATGTGTTTTTTATT............................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................AAGCGGGGATCTGGGTTTA.................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTATAAGTATGAAACG.................................................... | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................TTTTTTTTTTTTTTTTTTTTCT................................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................AAAGTGGCGATATGGGTTT................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AAAGCGGCAATATGGGTTT................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAAAGCGGGGATATGGGTTT................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:4027026-4027276 + | dvi_15484 | AAA----------A----CTGACGACGC-GTGGAGGTGAACT-----------------------GAGCTGAAGT-A----C--------CATAATAAATGGAATAAATGCGTGAAAATGTGTGCAC---------------------ACAAA-----TAAAAAAAAAAAAAAAAA--AAAA--------AGAAACCCGTACAC----------AAA-------------------AT----A-TCACTT---GAAGTT--------------GCGCCGCTAAACCCAAATGTTGACCCCAATTT------------------------TGTTT-GATTTTGATCGAATATTCAT-AT-TTTGCAGAGCCGGCACGAGAATGGTACGGGCGGCATTGGC---------------------------------------------------------------AAGAGCGC---CAGTGATGT--------- |
| droMoj3 | scaffold_6540:21025743-21025989 + | AAA----------A----CTGAC-GTTGCGCTTAGGTGAACT-----GA-AT---CCA---AC-GGAGCTGAAGT-A----C--------CATAATAAATGGAATAAATGCGTGAAAATGCGTGCGC--------------------------------CCAAAAATAGACCTAAATAGTAC----------A---CAAA------------------------------------AT----A-TCACTT---GAAGTT--------------GCGCCC-T--ATCGAATTGTTGACCCCGATTCTTGAATATCCAGTAAA-------------------TATATGCT----CAT-AT-TTTGCAGAGTCGGCACGAGAACGGTACGGACGGCT------------------------------------------------------------TTTTTGGCAAAAGCGA---TAGCGATGT--------- | |
| droGri2 | scaffold_14906:675495-675672 + | AAAAT----CTACT----CTGAC-GCTGCGTGGAGGTGAACTGA-----------------AC-TGTACTAAAGT-A----CTGTGTCTGTATAATAAATGGAATAAATGCGTGAAAATGTGTGCGC--------------------------------CCAAAAATAAATGCAGA--AAAA------------------------------------------------------AG----A-TCACTT---GAAGTTT-------------GCGCCC----AACAATATGTTGACCCCAATTT------------TGGATTT-------------------TTATT-TTATAT---------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004943:7929473-7929761 + | TAG----------A--------------------------------------------------CGAGGTGAAGC------G-------AGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCTGAT-------------C--------------CCAAAAATAAACCAAAA--ACAC----------AAACACAGACAACCAT------ACAC-----AACGAATACAGATGCACAAT-TCACATTAAGAAGTT--------------GCGCCGCC--AACGAAATGTTGACCCCAATTT------------TTCATCGT--CATCT----CC-TTTTTGGTT-TT---TGTT-TTTGCAGAGCCGCCACGAGAATGGAACTGGAGGAGGCGGAGGAGGAG------------GAGGAG---------------GCATAGAAGAAGGATGTATAAGTAAGACCGACGTTAATGATTT--------- | |
| dp5 | 2:26154731-26155034 + | AAC----------T----CTGCT----GCGTCTTCGTGACTTC----GC-ATCTGCCA---GC-AG----GGAGT-ACGATA-------CGATAATAAATGGAATAAATGCGTGAAAATGCT-GCCCCC-----------CAATCTGCCTCGAAAGTGTACAAAAATAAAC--AAC--GAA--------------------------A------GAAC-----------------TGC----TTTGTATT---GAAGTTTTTGCGAGGCGGCCCACCCG-----ACAAAATGTTGACCCCAATTTAA---CG-----TCAATTTT-------------------TGGT-TTTAATTTT-TTCGCAGAGCCGCCACGAGAATAGCAATGGAGGCGGAGAAG---CAG------------GAGGCACCTCTCCAACAGCTGTTGGCGGAGGG---------------------------GAGCTGGATGCAAC | |
| droPer2 | scaffold_6:1485361-1485664 + | AAC----------T----CTGCT----GCGTCTTCGTGTCTTC----GC-ATCTGCCA---GC-AG----GGAGT-ACGATA-------CGATAATAAATGGAATAAATGCGTGAAAATGCT-GCCCCC-----------CAATCTGCCTCGAGAGTGTACAAAAATAAAC--AAC--GAA--------------------------A------GAAC-----------------TGC----TTTGTATT---GAAGTTTTTGCGAGGCGGCCCAGCCG-----ACAAAATGTTGACCCCAATTTAA---CG-----TCAATTTT-------------------TGGT-TTTAATTTT-TTCGCAGAGCCGCCACGAGAATAGCCATGGAGGCGGAGAAG---CAG------------GAGGCACCTCTCCAACAGCTGTTGGCGGAGGG---------------------------GAGCTGGATGCAAC | |
| droAna3 | scaffold_13340:21659368-21659666 + | AACCGGCGCTG--A----CTGCCG-----------CCGTACT-----GTAG------TACGGCGGGCGTTGGAGTTT----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGATC-CGACGAT----CCACGCCGA-----TCCGAAAATAAACCCGCT--GCA---------------------GCCGCC------GAAC--------GGCT-CTGCTGC----T-TCAGTT---GAAGTT---GGGATGTG-CTGTGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CGTGTTTTGCT-TTGTTTGTT-TGCCATCTT-TTCGCAGAGCCGCCATGAGAACGGCAACGGAGGAGCTGGCG---AGG------------GCGGCG---------------CCGGAGGAGT------------------------------------------ | |
| droBip1 | scf7180000396413:879351-879663 - | CTG----------A----CTGCCGACTA---------------CACAGT------ATT---GC-GGCGTTGGAGT-T----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGATCTCCACGAT----CCACGC----------CGAAAATAAACCCGCT--GCAG----------------CCACA--GCCATCAGCGAGC-----------TGGTGGTGC----T-TCAGTT---GAAGTT--------------GTGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CGTGTTTTGCT-TTGTTTGTT-TGCCATCTT-TTCGCAGAGCCGCCATGAGAACGGCAACGGACTAGGCGGACTAGGAGGAGCCGGCGAGGGCGGTG---------------CAGGAGGAGCGGC---------------------CCATGATCTGGAGGC--- | |
| droKik1 | scf7180000302639:740779-741070 + | G-----------------CTGCCTTCTG---------------------------CCT---GC-GGTAGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTTT--CCCGAT-------------CCACGCCGAGA---TCCGTAAATAAACTCGAT--GTGC----------A---CCCCACACCGCC------TT----CGCTGCGGCTGCTGCTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAAATTT--CGTGTTT-TGT-TTGTTTGTC-TTCCATCTT-TTTGCAGAGCCGCCACGAGAATGGCAATGGAGTGGCAGGAG---GCGAA------TCAGG---------------------AGAAGGAGTGTC---------------------CCTAGAGCTGGAGGC--- | |
| droFic1 | scf7180000452523:1310936-1311218 - | AAC----------T----TTGTTGGA-------------------------------T---GC-GGCAGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGAT-------------CCGCGCCGA-----TCCGAAAATAAACTCGAA--GTAC----------A---CCCCACACCGCC---GGCGAACTCTGCTGCTGCTGCTGCCGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CGTGTTC-GCT-TTGTTTGTT-TTCCATCTTTTTCGCAGAGCCGCCACGAGAATGGCAATGGAGGCGGAGTCG---AAG---------GAGGAGGAG---------------CCGAAGGAGTG----------------------------------------- | |
| droEle1 | scf7180000491080:1716035-1716321 - | C------------------TGTCGGCTG---------------------------CCT---GC-GGCGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTTC--CCCGAT-------------CCACGCCGA-----TCCGAAAATAAACCCAAT--GTAC----------A---CCCCACACCGCC------GAAT--------------TGCTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CGTGT-T-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCACGAGAATGGCAATGGAGAAGTCGGAG---GAG---TCAGAGGAGGAGGTG---------------CCGAAGGAGTGGG---------------------CCACGATCTGGAGGC--- | |
| droRho1 | scf7180000779241:38692-38956 + | -------------------TGTTGGCTG---------------------------TCT---GC-GGCGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTTT--CCCGAT-------------CCACGCCGT-----TCCCAAAATAAACTCAAT--GTAC----------A---CCCCACACCGCC------GAAC--------------TGCTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--TGTGT-T-GCC-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCACGAGAATGGCAATGGAGGAGGG---------------------------G---------------TCGAAGGAGAGGG---------------------CCACGATCTGGAGGC--- | |
| droBia1 | scf7180000302136:1537432-1537703 - | CTG-C----CGATT----CCGTCGACTG---------------------------CCT---GC-GGCGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGAT-------------CCACGGCGA-----TCCGAAAATAAACCCAAT--GTAC----------A---CCCCACACCGCC------GAAA-----------TGCTGCTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTTCTCATGTTT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCACGAGAATGGCAATGCAGGAGGTGGAG---GAG------------TTGCAG---------------GCGAAGG--------------------------------------------- | |
| droTak1 | scf7180000415242:182879-183169 + | AAAC---------TCCGACGGCCGACTG---------------------------CCTGCGGC-GGTGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGAT-------------CCACGGCGA-----TCCGAAAATAAATTCAAC-AGTAC----------A---CCCCACAC----------------------------TGCTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CATGTTT-GCT-TTGTTTGTTTTTCCATCTT-TTCGCAGAGCCGCCACGAGAATGGCAACGCAGGAGGAGGAG---GAGGA------GCAGGAGGAG---------------CTGAAGGAGTGGG---------------------CCACGATCTGGAGGC--- | |
| droEug1 | scf7180000409797:1614815-1615097 + | CTG----------G----CTGCCGGTTG---------------------------CCT---GC-GGCGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTTC--CCCGAT-------------CCACGCCGA-----TCCGAAAATAAACTCAAT--GTAC----------A---CCCCACACTGCC------GAAC--------------TGCTGC----T-TCAGTT---GAAGTT--------------GTGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TCAATTTT--CGTGTTT-GCT-TTGTTTGTT-TTCCATCTT-TTTGCAGAGCCGCCATGAGAATGGCAATGGAGGAGGAGTAG---AAG------------GAGGAG---------------CCCAAGGAGTGGG---------------------CCATGATCTGGAGGC--- | |
| dm3 | chr3R:11916748-11917017 - | G------------------------CTG---------------------------CTG---AC-GGCGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGAA-------------CCACGCCGA-----TCCGAAAATAAACTCAAT--GTAC----------ATGCCCCCACACCGCC------GAAC--------------TACTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TTAATTTT--CGTGTCT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCATGAGAATGGCAATGGCGGAGGAGAAG---AGG------------GAGGAG---------------CCAAAGGAGTGGG---------------------CCACGATCT--------- | |
| droSim2 | 3r:9334067-9334346 + | AAC----------T----CTGCAGGCTG---------------------------CTG---GC-GGTGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGC--CCCGAA-------------CCACGCCGA-----TCCGAAAATAAACTCAAT--GTAC----------ATGCCCCCACACCGCC------GAAC--------------TACTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TTAATTTT--CGTGTCT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCATGAGAATGGCAATGCCGGAGGAGAAG---AGG------------GAGGAG---------------CCAAAGGAGTGGG---------------------CCACGATCT--------- | |
| droSec2 | scaffold_0:10042620-10042899 + | AAC----------T----CTGCAGGCTG---------------------------CTG---GC-GGTGGTGGAGT-A----G-------GGATAATAAATGGAATAAATGCGTGAAAATGTGA--CCCGAA-------------CCACGCCGA-----TCCGAAAATAAACTCAAT--GTAC----------ATGCCCCCACACCGCC------GAAC--------------TACTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TTAATTTT--CGTGTCT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCATGAGAATGGCAATGCCGGAGGAGAAG---AGG------------GAGGAG---------------CCAAAGGAGTGGG---------------------CCACGATCT--------- | |
| droYak3 | 3R:14517635-14517921 + | CAG----------G----CTGCTGG------------------------------CCT---GC-GGCGGTGGAGT-A----G-------GGATAATATATGGAATAAATGCGTGAAAATGTGC--CCCGAT-------------CCACGCCGA-----TCCGAAAATAAACTCAAT--GTACCCCGCACTCAATACCCGCACACCGCC------GAAC--------------TACTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTAA---C------TTAATTTT--CGTTTCT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCATGAGAATGGCAATGGCGGAGGAGAAG---AGG------------AAGGAG---------------CCCAAGGAGTGGT---------------------CCACGATCT--------- | |
| droEre2 | scaffold_4770:11440287-11440572 - | AAC----------T----CTGCAGGCTG---------------------------CTG---GC-GGTGGTGGAGT-A----G-------GGATAATATATGGAATAAATGCGTGAAAATGTGC--CCCGAT-------------CCACGCCGA-----TCCGAAAATAAACGCAAT--GTAC----------ATACCCGCACACCGCC------GAAC--------------TACTGC----T-TCAGTT---GAAGTT--------------GCGCCG-----ACAAAATGTTGACCCCAATTTGA---C------TTAATTTT--CGTGTTT-GCT-TTGTTTGTT-TTCCATCTT-TTCGCAGAGCCGCCATGAGAATGGCAATGGCGGAGGAGAAG---AGG------------AAGGAG---------------CCAAAGGAGTGGG---------------------CCACGATCTGGAGGT--- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:30 AM