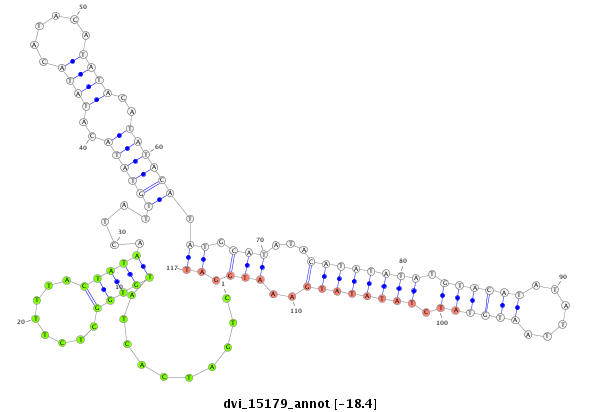
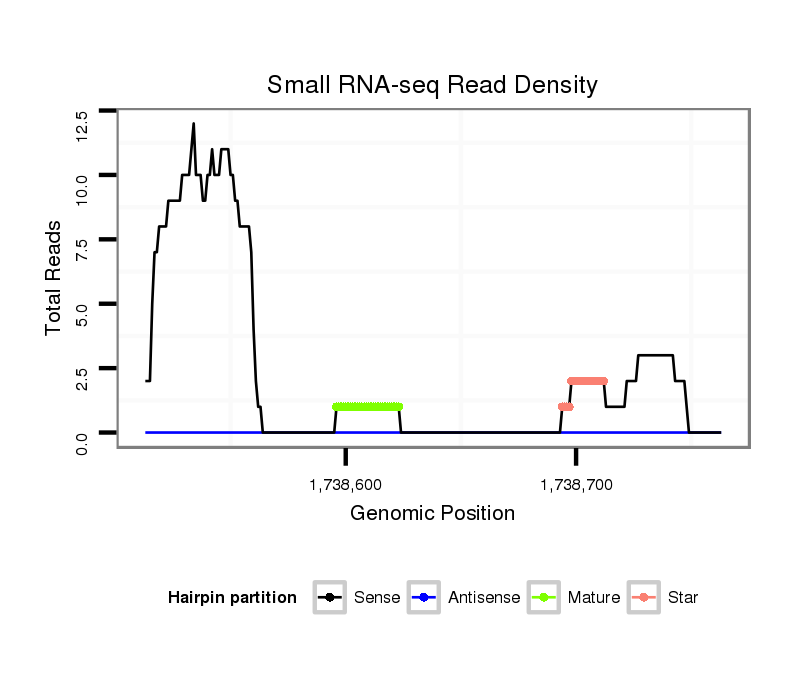
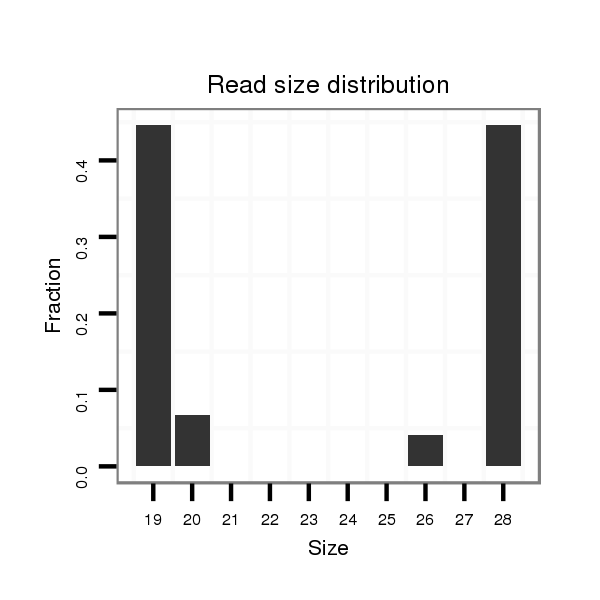
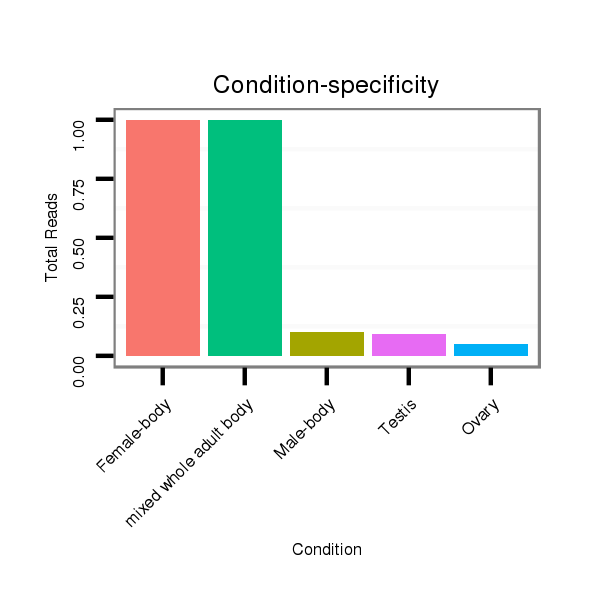
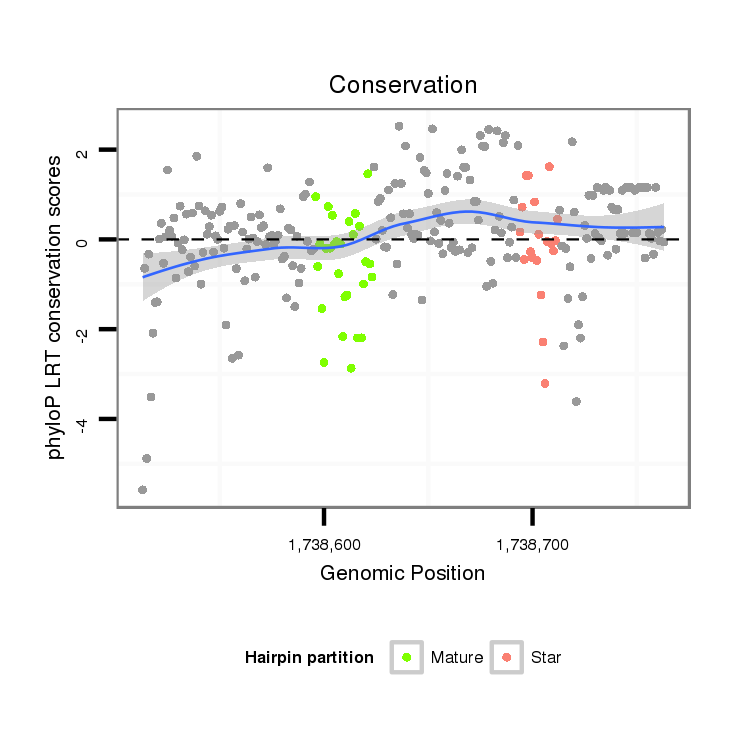
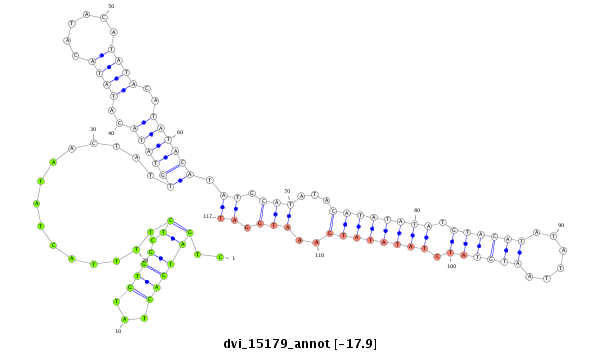
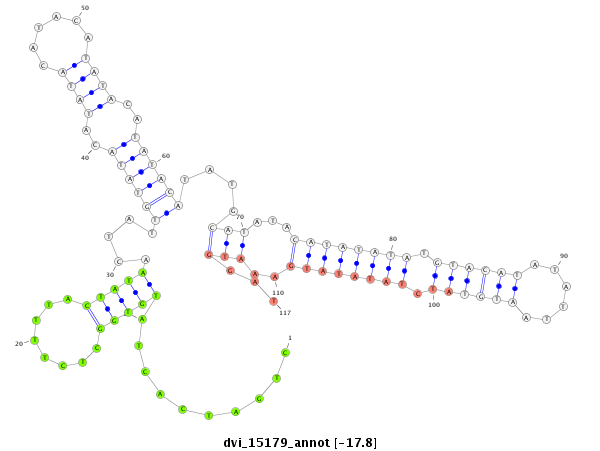
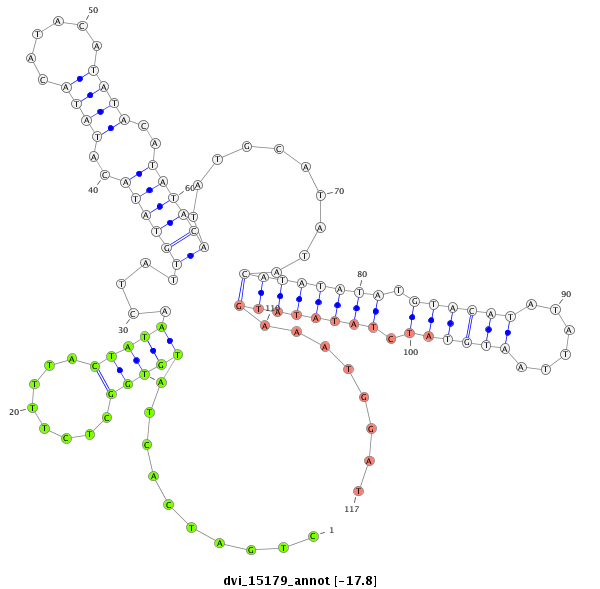
ID:dvi_15179 |
Coordinate:scaffold_13047:1738563-1738713 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -17.9 | -17.8 | -17.8 |
 |
 |
 |
exon [dvir_GLEANR_9052:7]; CDS [Dvir\GJ23721-cds]; intron [Dvir\GJ23721-in]
| Name | Class | Family | Strand |
| (CATATA)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTAAGGGCTATACCGATGAGGGTGAATTTGACTGGACAGGGAAGACCATGGTAGGTAACAAGTTAAAACGTAAAATCGATACGCTGATCACTATGTGGCTCTTTTACTATAACTATTGTATACATATACATACATATACATATACATATGCATATACATATATATGTACATATATTAATGTATCTATATATGAAATGGATATTCGCATTCTCATTGACAATCACCTGAGCCGAATTCTAAACTCACATTTA ***********************************************************************************..........(((((........))))).....((((((..((((......))))..)))))).((.(((...((((((((.((((((......)))))).))))))))..))).))*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060657 140_testes_total |
M047 female body |
SRR060663 160_0-2h_embryos_total |
SRR060678 9x140_testes_total |
SRR060664 9_males_carcasses_total |
SRR060660 Argentina_ovaries_total |
M061 embryo |
SRR060667 160_females_carcasses_total |
SRR060670 9_testes_total |
SRR060679 140x9_testes_total |
V116 male body |
SRR060685 9xArg_0-2h_embryos_total |
M027 male body |
SRR060656 9x160_ovaries_total |
SRR060658 140_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060668 160x9_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR1106729 mixed whole adult body |
V047 embryo |
SRR060681 Argx9_testes_total |
SRR060669 160x9_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...AGGGCTATACCGATGAGGG..................................................................................................................................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GGACAGGGAAGACCATGGA....................................................................................................................................................................................................... | 19 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TTGACTGGACAGGGAAGACCATGT........................................................................................................................................................................................................ | 24 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GGACAGGGAAGACCATGT........................................................................................................................................................................................................ | 18 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................TGACTGGACAGGGAAGACCATGT........................................................................................................................................................................................................ | 23 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TACCGATGAGGGTGAATTTG............................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GGGCTATACCGATGAGGGTG................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AGGGCTATACCGATGAGG...................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GCTATACCGATGAGGGTGAAT................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGACAATCACCTGAGCCGAATT............... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................CTGATCACTATGTGGCTCTTTTACTATA............................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GGGCTATACCGATGAGGGTGAAT................................................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GTAAGGGCTATACCGATGAGGGTGA.................................................................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GGACAGGGAAGACCATGG........................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TTGACTGGACAGGGAAGACCA........................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ATTTGACTGGACAGGGAAGACCATGT........................................................................................................................................................................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GACTGGACAGGGAAGACC............................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TTGACTGGACAGGGAAGACC............................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TGAGGGTGAATTTGACTGGAC...................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AATTTGACTGGACAGGGAAGACCA........................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GTAAGGGCTATACCGATGAGGGT.................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GGACAGGGAAGACCATGTC....................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................TGGACAGGGATGACCATG......................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TTGACTGGACAGGGAAGACCAT.......................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CATTGACAATCACCTGAGCCGAAT................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GACTGGACAGGGAAGACCATGT........................................................................................................................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................GAATTTGACTGGACAGGGAAGAC............................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GTGAATTTGACTGGACAGGGAAGACC............................................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GTGAATTTGACTGGACAG.................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ATCTATATATGAAATGGAT................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GGTGAATTTGACTGGACAGGG.................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CTCATTGACAATCACCTGAGC..................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................AGCCGAATTCTGAACTCACATTC. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ATATATGAAATGGATATTCGCATTCT........................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............ATAACGGTGAATTTGACTCG........................................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ATACATATACATATACATAT...................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TATACATATACATATGCATATACATA........................................................................................... | 26 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ATACATATACACATACATATGC.................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................ATTGTATGGATATACAGAC...................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TACATAAGCGTATACATATATAT...................................................................................... | 23 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................ACGATGATCACTAGGTGTC........................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................AATACATAAGCGTATACATATATAT...................................................................................... | 25 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................TATACATATACATACATATA................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CATTCCCGATATGGCTACTCCCACTTAAACTGACCTGTCCCTTCTGGTACCATCCATTGTTCAATTTTGCATTTTAGCTATGCGACTAGTGATACACCGAGAAAATGATATTGATAACATATGTATATGTATGTATATGTATATGTATACGTATATGTATATATACATGTATATAATTACATAGATATATACTTTACCTATAAGCGTAAGAGTAACTGTTAGTGGACTCGGCTTAAGATTTGAGTGTAAAT
***************************************************..........(((((........))))).....((((((..((((......))))..)))))).((.(((...((((((((.((((((......)))))).))))))))..))).))*********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060673 9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................ACCTATGCTACTAGTGAAACACC......................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TGTATACGTATATGTATATATA...................................................................................... | 22 | 0 | 9 | 0.56 | 5 | 0 | 3 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................ACGTATATGTATATATACAT................................................................................... | 20 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................TGAAAGCGTAAGAATAACTGT................................ | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................GTATACGTATATGTATATATA...................................................................................... | 21 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:1738513-1738763 + | dvi_15179 | GTAAGGGCTATACCGATGAGGGT-GAATTTGACTGGACAGGGAAGACC-ATGGTAGGT-----------------AAC-AAGTTAAAACGTAAAATCGATAC-------GCTGATCACTATGT-------------------------------------------------------------G----------------------------------------------------------------GCTCTTT-----------TACTATAA--------------------------------------------CTATTGTATACATA----TACA--TA--------------------CATATACA----T----------A------TACATATGCA--T----ATACATAT--ATAT--GTAC----------------ATA--------TA-----------TT----AATGTAT--CTATATA--TGA-----A---------A-----------------TGGAT---------AT-TCGCATTC------------------TCAT---------TGACAA---TCACCTG--AGCCGAATTCTAAACTCACATTTA |
| droMoj3 | scaffold_6540:15441116-15441310 - | GTAAGGGCTATACCGACGAGGGC-GAATTTGACTGGACAGGGAAAACC-ATGGTGGGT-----------------AGC-AAGTTGAAATTTATTAC-----C-------------------G------------------AT--------------------TAACAATGA----T------------------------------------AATCACCG-----------------------------------------------------------------CAA------------------------GATCC-A-TATTGAAAATATG----TGCA--------------------------------------------------------T---------------------------------------------------------------------------------------------ATT--TGA-----A---------A-----------------TTGAT---------AATTCACATTT------------------GCCA----TT---TAACAAAATTCATCTGCGAGCCGAATTCTAAACTC--ATCTA | |
| droGri2 | scaffold_14906:12554792-12555003 + | GTAAGGGCTACACCGACGAGGGC-GAGTTCGACTGGACAGGGAAGACA-ATGGTAGGT-----------------AACCCAGTTGAAAC------------T-------------------G------------------AT--------------------GAAC---AGACAAA------------------------------------AATCAACGCGTCTCGATGCTATTATTATTATTGT-------------------TATT--ATTA------------------------------------------------CG--------ATAA--C----A------------------TACAA----A--CACACACA--------------T---------------------------------------------------------------------------------------------TTG--TCA-----A---------A-----------------TGGAT---------AA-TCACATTC------------------GAAA----T-------CA---TGCATCTG--TGCCGAATTCTAAATTCATATCTA | |
| droWil2 | scf2_1100000004943:5553101-5553285 - | GTAAAAAAAATACAAAATATGCA-TAATTAAAAGGAAAAATGCAAAAC-GGCGTAGTT-----------------AAT-AAAACGCGAGTGCATCA----AT-------A------------------------------A-TTGCCAT--------------------AATCT----------------------------------------------------------------------------------------------------------CTCCTCAA------------------------CATAC-A-TA-TATATAAATA----TTC--------------------------------ATACATATATATATG----------TA--------TATAT----------ATAT--ATATATACATA----TATATATA--------TA-----------TA----CATATAT--A------------------------------------------------------------------TTT------------------ATA---------------------------------------------------A | |
| dp5 | XL_group3b:258285-258465 + | AGGGCG------------AAAGT----TTCGTCTGAAAAGGAGCGCCCAATAATAAAAAGGAAGTTTTTATCAGAA------------AGTCAAATGGAAAT-------G------------------------------C-ATGGCGT--------------------GAGTCAC---------AC---------------------------TCTGC------------------------------------------------------------------------G--------------------TATAT----C-TATGTACATA----TAC--------------------------------ATACATATACATATA----------CA--------TATAT----------TTAT--CTTTACATATA----CATACATA--------TA-----------TT----TATATTT--A------------------------------------------------------------------TAT------------------TTA---------------------------------------------------T | |
| droPer2 | scaffold_21:1464490-1464670 + | TTAACT----------------------------TGAACGGGAAAACA-ATTAAAGCT-----------------AAT-GAAA-GGAGAGTAAAAT-----TGCAGC--GCGAAA--GAGTTG------------------------------------------------------------------------------------------------------------------------TAGC--AGGAATC--------TAATAAA--------------------------------------------ACAAATATTATATATATA----TATA--TA--------------------TATATATA----T----------A------TATATATATA--C----ATACAT----ATA------------------TA----TA----------TGATA-T---ATATACAACTATAT--ATATTTT--T-----------------------------------------------------CAAAAGT------------------ATG---------------------------------------------------T | |
| droAna3 | scaffold_12903:223608-223752 + | CAAATGCAA------------------------------------------------------------------------------CAACAA---------------------------------------------------AACCA--------------------------ACATCACTGAGCTTTGTA-----------------------------------TGTA-------------------TTTCTGAGCGCTCTAACCAA--------------------------------------------AC-----------ATATA------CATAAA--------------------TACAT--A--TAT----------A--------AATATGTATAT----ATA--TAT--ATATGTGTGT------A----T----GTG--------TA-----------TA----TATAT---------------------AATATATGTAG-----------------TGGAA----------------------------------------A--------------------------------------------------CT | |
| droBip1 | scf7180000395977:9132-9303 - | ATGAGAGCTATAC--ATGAAAGC-TATTTT---T-----GGCAAAATA-ATAT------------------------------------------------TAC---GT----AC--CAAATTTATAGGGATC------------------------------------------------------------------------GGCC--------------------------G---------ACTATATCTTA-----------T---ATAA-----------TAAT---------------------------CT------TACATATA------CATATA--------------------TATAT--A--TAT----------ATATATATATA--------T----ATA--TAT--ATATATATA------------------TA--------TAGTACAATATAAT------------------AAA--TAT-----A------------------------------------------------ATAC------------------ATA---------------------------------------------------T | |
| droKik1 | scf7180000302696:811756-811931 - | AACAAATTTAA----------------------------------------------------------------------------CATTAT-------------------------------------------ATGC--------A--------------------------TGCTCGTTACGTTTGACTTGATTTGGATTTTGCTAGCTGCAA-------------------------------------------------------ATAT-------------CAA--------------------TATAT----A-TATATACATA----TAC--------------------------------ATACATATATATATA----------TA----------TCAA----T----AT----ATATATATACATATACA----TACATACATATA-----------TA----TATATAT--A----------TATATAT---------------------------------------CAAT-ATCAATCA--------------------A-------------------------------------------------TTG | |
| droFic1 | scf7180000454040:238146-238283 + | GTAAGGTCGTTT-----------------------------------------------------------------------------------------CGC---TC----CC--CAATAT------------------------------------------------------------------------------------------------------------TGTTG---------GCCTTTTCTTC-----------T---ACTA-----------TAGG---------------------------AA------TACATATA------TGCATG--------------------TAC----ATATGT----------ATACATACGTC--------C----ATAAAC----GTATATGTA------------CA----TA----------TTATC-T---ATA------------------------AAAAAAT-----------------------------------------GT-TTACATCC------------------ACA-------------------------------------------------TTG | |
| droEle1 | scf7180000491193:54336-54515 - | GAAAACTCTGG---------------------CG---------------------GCC-----------------A-T-AA--AAAAGGATAAAAGCGAAAC-------ACAGGGTACAATGT-------------------------------------------------------------G-----------------------------------------------GCTG---------ATG----CCTT-----------T--------------GGGTTAAG----------------------CACAT-A-TA-TATACACATA----TAT--------------------------------A--TACAGATATATA----------TA--------TATATA----T----AT----ATCTATATACA----ACTAGATA--------TA-----------TC----TGTGTGT--GTATGTG--CCTCAAG-G---------------------------------------ACAT-TTCCACTT------------------GCA---------------------------------------------------T | |
| droRho1 | scf7180000776638:16511-16639 + | ATTATATATAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTA--------TAAT----A--T-------------ACATATACATATACATATACATATACAT-A-TA-CA--------TATA------------------------------------------------------------C----------ATATA----C--AT------ATACATATACATATACA----TATACATATAC-------------------------ATATACATATA--CATATAC---------------------------------------ATAT-ACATATAC------------------ATA---------------------------------------------------T | |
| droBia1 | scf7180000298858:2780-2915 + | AAATTA-------------------------------------------------------------------------------AAAAATATCA------------------------------------------------------------------------------------------------------------------------------------ATAAT-TTATTATTATTATT--ATTATTA--------TAATATA--------------------------------------------ACATCTACTATATATATA----TATA--TA--------------------TATATATA----T----------A------TATATACACA--T----ATATAC----ATATATGTACGC----A----TATA------------C-----------AT------------------ATA--TAA-----A---------A-----------------TAAAT----------------------------------------A--------------------------------------------------TT | |
| droTak1 | scf7180000411343:21714-21840 - | TATATGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATATATATATGTAT--------------------------------ATATAT----ATATG----------TA--------TATAT----------ATAT--ATGTATATATA----TATATGTA--------TA-----------TA----TATATATGTATATATA--CATATAT-GATTTTTATAGCAAACGTTTGAAAAGCATACTT----------------------------------------G--------------------------------------------------TT | |
| droEug1 | scf7180000409752:791160-791336 - | CTGCTG------------AGAGT-AAT-TTCAATGGTAAGTCAAAAAC-ATAT------------------------------------------------CTT---GT----AT--TA-------------------------AATCGGAGGTAAAGAAAGCAAT---AAACTAA------------------------------------ACTAAATTGGTTT------------------------------------------------------------CG----A--------------------TGTAT-T-TA-TTTTCATATA----TAT--------------------------------A--TATATATATATATG--------TA--------TATAT----------ATAT--ATATATATATA----TATATATA--------TA-----------TG----TATTTAT--T------------------------------------------------------------------TAT------------------ATA---------------------------------------------------T | |
| dm3 | chrU:6539654-6539855 - | CAGACGGCTAAGCCAGATGACGCTGGCTAGAACT-AGC----ACGATA------------------------------------------------------AG---GA----AG--TAGCTGAAGAGGGATC-----------CCAAT--------------------AGGAA-----------CCTTTATA-------------------TATGT-----------------------------------AT------------AT----A--T-------------GTATATATGTATATATGTATATATGTAT-A-TA-TG--------TATA------------------------------------------------------------T----------ATGTA----T--AT------ATGTATATATGTATATA----TGTATATATGT-------------------------ATATATGTATA--TATGTAT---------------------------------------ATAT-GTATATAT------------------GTA---------------------------------------------------T | |
| droSim2 | x:14968001-14968158 + | CAAGTCA------------------------------------------------------------------------------AAAAGTATAT------TTT---CT----TT--TA-------------------------AATTG--------------------------ACATTAAACC----------------------------------------------------------------GCTTTTT-----------TGCCACAA--------------------------------------------CAACCCTAAATATA------TCTATA--------------------TACAT--A--TAC----------A------CATATATATA--A----ATATAT----ATATATGTAA------A----AA----TA--------T-----------AT------------------ATA--CGA-----A---------A-----------------TACGTGTTTCCAATAT-TTTTATGT------------------ACA--------------------------------------------------AT | |
| droSec2 | scaffold_0:16589188-16589327 + | AATGAGCTTAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCAAACAAACAGCAACATTAAATTAA-----------------------------------------------GT----A--TACATCTCCATTA------------------------TATAT-A-TA-TATATATATA----TAT--------------------------------ATATACATATGTATA----------TA--------TATAT----------ATAT--ATATATATATA----TATACATA--------TA-----------TA----TATATAT--A------------------------------------------------------------------TAT------------------ATA---------------------------------------------------T | |
| droYak3 | 3L:19816986-19817219 + | AATTTG-------------------------ACTGAAAAGAGAATACA-ACGGAAA--------ATTAGAGTACAC-------------------------T-------------------GT----------CGTATGC----AGAAA--------------------------TATTTGTTGTGCCTTGCA-----------------------------------TATTATTATTATTTTTTT-------------------TACC--AATA-----------------------------------------------------TTAAG------TCTTAGCTGACTTGAGTAAATATATACATA------CATATATG--------------CATGCA----A----------------------------------------ATA--------TA-------------------ATTAC--T---------------------------------------------------------GAA-G---ATCAATTAAATTGCTCGATTTTGAAAATCTTCGAATAATAG---TCACC------TCAAATACAAAAATTGTACTTA | |
| droEre2 | scaffold_4929:21197034-21197192 - | TAATAATT-------------------------------GGTAAATTT--------------------------------------------ATA-------------------------------------------------------------------------------------------------TCGATATACATATCGATATTTATTTAC-----------------------------------------------AT----A--T-------------ACATATACATATACATATACATATACAT-A-TA-CA--------TATA------------------------------------------------------------C----------ATATA----C--AT------ATACATATACATATACA----TATACATATAC-------------------------ATATACATATA--CATATAC---------------------------------------ATAT-ACATATAC------------------ATA---------------------------------------------------T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:46 PM