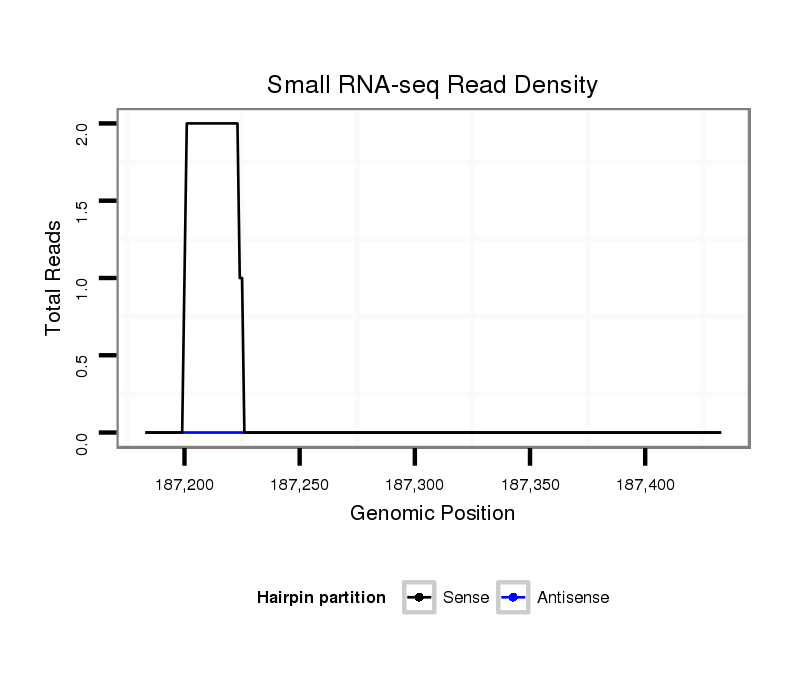
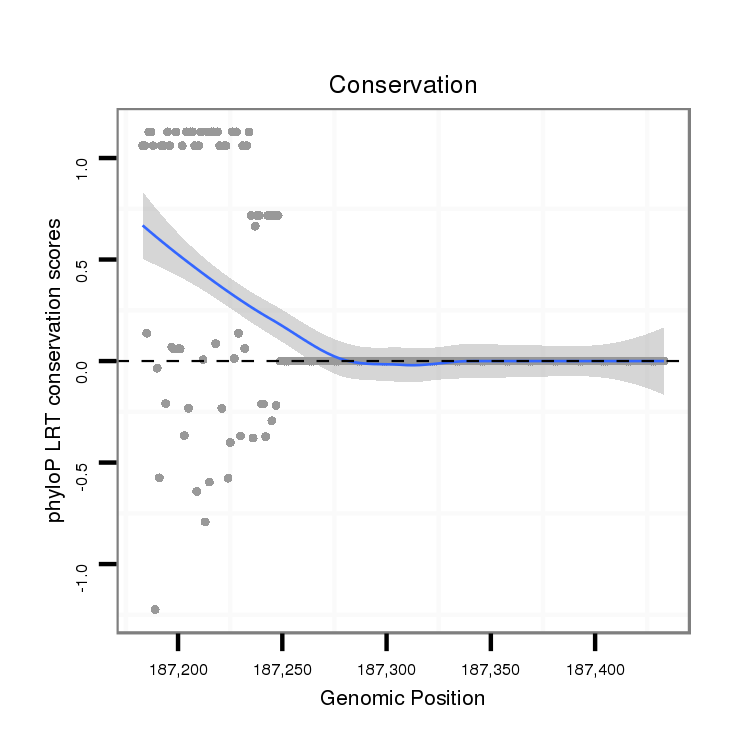
ID:dvi_14859 |
Coordinate:scaffold_13047:187233-187383 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_8952:6]; CDS [Dvir\GJ23613-cds]; intron [Dvir\GJ23613-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGAATGTGTGGGACAATTTCAATATCCGACCTCAACACAGCTTTGAATGAGTAAGTATACAACACATACAAAATATGTTGCAACAAGATAAGCATCTAGCAACAAAAAATACATAATATACGTGTCCAATAAGCTATGAAACATCTATGGATAAACAACTAAGTTATTAAAGTCGAAAACTTGTTTATCCACCGATTTTATGGAGCTATAACTTGTTCGACGTGTATATAAAATGCCTTATAATAACAAAA **************************************************.........................((((((....((((....)))).))))))......((((.......)))).................(((..((((((((((....((((.((......)).))))))))))))))..))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
GSM1528803 follicle cells |
M047 female body |
V053 head |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
M027 male body |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................AATACCGTATAATAACAAA. | 19 | 2 | 7 | 1.86 | 13 | 6 | 0 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 0 |
| ..................TCAATATCCGACCTCAACACAGCTT................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TTCAATATCCGACCTCAACACAGC.................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............AAATTTCAATATCCGACCTC.......................................................................................................................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TGGGACAATTTTAATATCTGA.............................................................................................................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TAATACCGTATAATAACAAA. | 20 | 3 | 20 | 0.35 | 7 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................................AATACCGTATAATAACAAAA | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTTATGGAGCTATAACTCT.................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................ATACCGTATAATAACAAA. | 18 | 2 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TAATACCGTATAATAACAAAA | 21 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................AACCTCTGTGGATAAAGAA............................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CCTTACACACCCTGTTAAAGTTATAGGCTGGAGTTGTGTCGAAACTTACTCATTCATATGTTGTGTATGTTTTATACAACGTTGTTCTATTCGTAGATCGTTGTTTTTTATGTATTATATGCACAGGTTATTCGATACTTTGTAGATACCTATTTGTTGATTCAATAATTTCAGCTTTTGAACAAATAGGTGGCTAAAATACCTCGATATTGAACAAGCTGCACATATATTTTACGGAATATTATTGTTTT
**************************************************.........................((((((....((((....)))).))))))......((((.......)))).................(((..((((((((((....((((.((......)).))))))))))))))..))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
V053 head |
SRR060672 9x160_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................TTTCAGTGTTTGAACAAGTAGG............................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGGTTGTTCGATACTTTGT............................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...TACAGACCCTGTTAAAGGTAC................................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................GTAGATGCCTATTTGGTTA........................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13047:187183-187433 + | dvi_14859 | GGAATGTGTGGGACAATTTCAATATCCGACCTCAACACAGCTTTGAATGAGTAAGTATACA-A--CACATACAAAATATGTTGCAACAAGATAAGCATCTAGCAACAAAAAATACATAATATACGTGTCCAATAAGCTATGAAACATCTATGGATAAACAACTAAGTTATTAAAGTCGAAAACTTGTTTATCCACCGATTTTATGGAGCTATAACTTGTTCGACGTGTATATAAAATGCCTTATAATAACAAAA |
| droMoj3 | scaffold_6540:1256263-1256331 + | GGAATGGGCGGGACTATTTCAATATCCGAGATAAATACAGCTATAAACGGGTAAGTATATATATACATA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14906:13713644-13713709 - | GGAATGACTGGAACAATTTCCAAATCAGAGATCAACACTGCGTTAAACGGGTACGTACGTA-A--AACA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_12916:8491136-8491187 + | GGGATGCGGGGAACACTGGCCAAATCGGAGCTAAACACTGCCATAAGTGGGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 05/16/2015 at 11:55 PM