




ID:dvi_14681 |
Coordinate:scaffold_13042:4118160-4118310 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.8 | -18.8 | -18.8 | -18.3 |
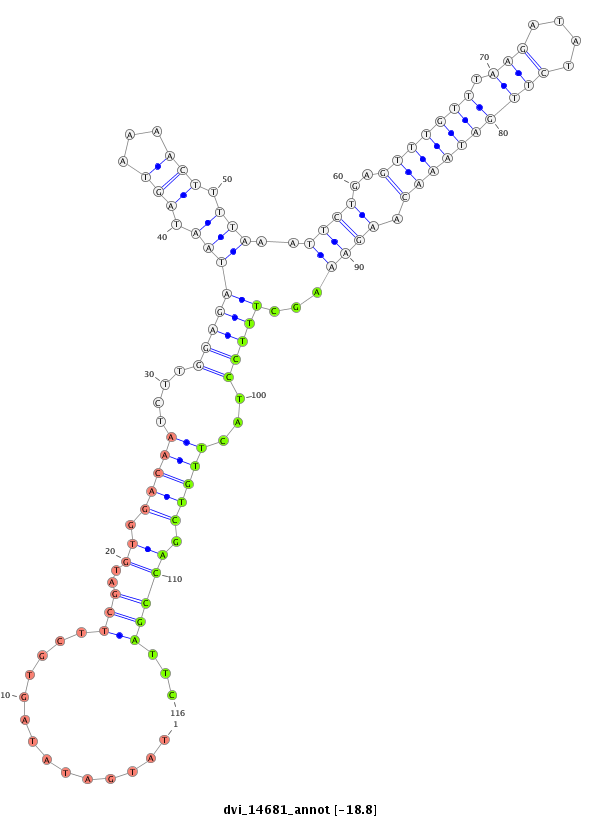 |
 |
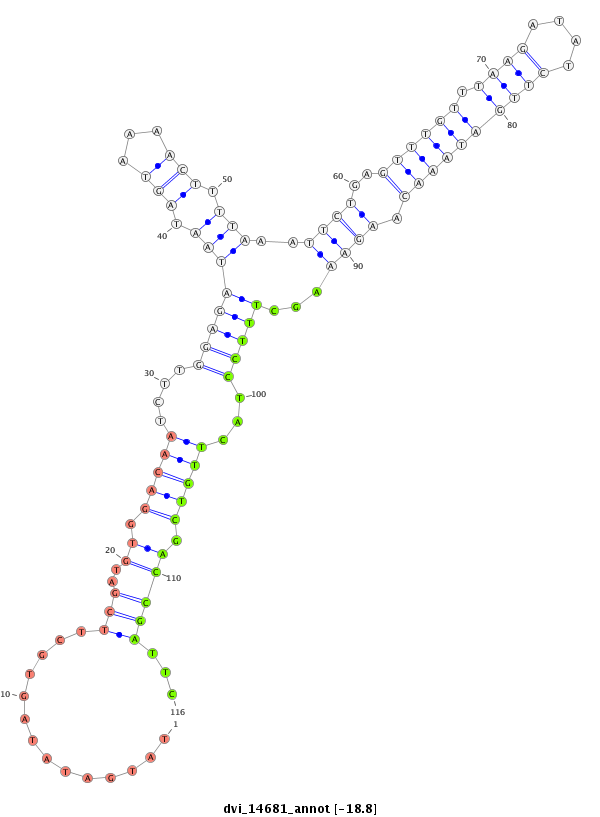 |
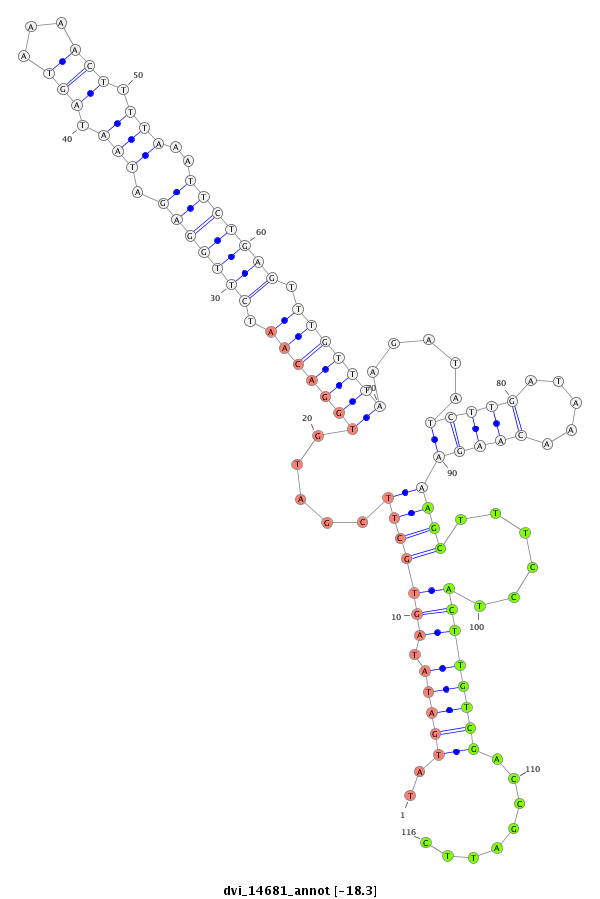 |
CDS [Dvir\GJ15991-cds]; exon [dvir_GLEANR_16415:2]; intron [Dvir\GJ15991-in]
| Name | Class | Family | Strand |
| Helitron-2N1_DVir | RC | Helitron | - |
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GAGCTATAGAGCTAAGGCAGTGTTAGGACTCGGCTCAGATGTAGGCATCCTTCCTATTGCAGCTATATGATATAGTGCTTCGATGTGGACAATCTTGGAGATAATAGTAAAACTTTTAAATTCTGAGTTTGTTTAAGATATCTTGATAAACAAGAAAGCTTTCCTACTTGTCGACCGATTCCTTTTATGTATATTATATAGTCGTCCGATCCGGCAGACATATGGCCTGCATCGGAAAGATGGACCCAAGC *****************************************************************...((..............((.(((((....(((((....................(((..(((((.(((((....)))))))))).)))....)))))...))))).))....))********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M028 head |
SRR060689 160x9_testes_total |
V116 male body |
SRR060681 Argx9_testes_total |
M047 female body |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................AGCTTTCCTACTTGTCGACCGATTC...................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AGCTTTCCTACTTGTCGAACGATTC...................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................AGTTCTTCGATGTGGACACTCT............................................................................................................................................................ | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TATGATATAGTGCTTCGATGTGGACAA............................................................................................................................................................... | 27 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................AGGACTGGGCTGAGATGT................................................................................................................................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................GTTCTTCGATGTGGACACT.............................................................................................................................................................. | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GGGAAATCTTGGAGATAATC................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................CAAGGAAGCCTTCCAACTT.................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CTCGATATCTCGATTCCGTCACAATCCTGAGCCGAGTCTACATCCGTAGGAAGGATAACGTCGATATACTATATCACGAAGCTACACCTGTTAGAACCTCTATTATCATTTTGAAAATTTAAGACTCAAACAAATTCTATAGAACTATTTGTTCTTTCGAAAGGATGAACAGCTGGCTAAGGAAAATACATATAATATATCAGCAGGCTAGGCCGTCTGTATACCGGACGTAGCCTTTCTACCTGGGTTCG
**********************************************************************...((..............((.(((((....(((((....................(((..(((((.(((((....)))))))))).)))....)))))...))))).))....))***************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060678 9x140_testes_total |
SRR060659 Argentina_testes_total |
V047 embryo |
V053 head |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................TCTACATCCGTAGGAAGGATAACGT.............................................................................................................................................................................................. | 25 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................GTAGGAAGGATAACGTCGATA......................................................................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGTACAACCTCTATCATCA............................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................GAGGTAGGATAACGTCAATATACTAT................................................................................................................................................................................... | 26 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................GGATAACGTCGATATACTAT................................................................................................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CTGAAGATTTAAGACTCA........................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................GGTAGGATAACGTCAATATACTATA.................................................................................................................................................................................. | 25 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AGGATAACGTCGATATACTA.................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:4118110-4118360 + | dvi_14681 | GAGCTATAGAGC---TAA-------GGCAGTGTTAGGA-----------------------CTCGGCTCAGATGTAG--GCATCCTTCCTATTGCAGCTATATGATATAGTGCTTCGATGTGGACAATCTTG--------------------------------------GAGATA--A-TAGTAAAACTTTT----------AAATTCTGAGTTTGTTTAAGATA------------------------TCTTGATAAACAAGAAAGCT----------TTCCTAC------------TTGTCGACCGATTCCTTTTAT-----GTATATTATATAGTCGTCCGATCCGGCA----------GACATATGG-CCTGCATCGGAAA----GATGGACCCAAGC |
| droGri2 | scaffold_15203:8268556-8268731 - | AT---------------------------------------------------------------------------------TGTTCCTATGGCAGCTATATAATATAGTAGTCCGATGTGACTGATTTTT------------------------------------TGTCCATGTGTG---CAGCACATAATTGAACGCATATTCATTAAGTTTGGTTCGAATA------------------------TTTTGTTA--------------------------CAAGAACTC------ACTTTCACCGATCATTCCTATGGCATCTATATGATACAGTTATCCGATCCGAC--------------------------------------------------- | |
| droWil2 | scf2_1100000004866:27812-28048 - | AG-TTATTGACA---AAA-------GTCACTG---------------------------------------TTTTCGACCGATCGTTCCTCTGGGAGCTATATGATATAGTCACCCGACCTTGATCAAATTT------------------------------------GGCAGAGGCGTATAAT-AAACTCTC----------CAATATTAAATTTCATGAGAATA------------------------GCTCAGAAAATAACGAAGTT----------ATTGAGAAAAGTCACTG-TTAGTGACATTGCCGTTTGTATGGGAGGTATATGATATAGTGGTCCGATCCGGCT----------GATATAGTGGTCTACA------------------------ | |
| droAna3 | scaffold_13417:3829579-3829785 - | --------------------------------------------------------------------------------------TTCTATGACGGCTATAGGATATAGTCGGCAGATCCTTATACAATTT------------------------------------TTCAGATA--A-GAT-----ATCTT----AAATTTAACTTGTGTGCAAAGTCCCTCTA------------------------GCTCGAAAAACACCAAAGTT----------ATGAC-------------ATTTCCGATCAATCAGTTATATGGCATATATATGATATAGTCGACCGATCCGACT----------TATATACTA-CCTGCAAGGAAAA----AAAGGATGTGTGC | |
| droBip1 | scf7180000395214:8332-8531 - | AT---------------------------------------------------------------------------------TTTTGCTATGACAGCTGTATGATATAGTTTTCCGATTTGGACTATATTT------------------------------------GGCAA--------------------------GCATAGTTTGTATGTTAGGTTTACATA------------------------TCTTGAAAAACAAAAAATTT----------TTTCCTACTATATACTT-ATTTCTATCCTATCGTTCCTATGGCAGCTATATGATATAGTCGTACGATATGGCC----------AATTTTTTT-ACTGCATTTCAAA----TAT---------- | |
| droKik1 | scf7180000301587:13299-13604 - | GTTC-------C---TAT-------GGCAGCTATAAGATATAGTCGTCCGATTTACATAAAATTTATACAAAAATTC--CGATCATTCCTATGGCAGCTATAAGATATAGTGGTCCGATTTTCATAAAATTTTAAACAAAATTCTGGAATAATATAAAATGGCTATATCTCAAAGATGA-TG--------------------------------------AAATAA------------------------TATTGAAAAACAGCCAAGTTATAATTTTTTT-TCCAA------------AAATTTATCGAACATTTGTATGGCAGCTATATGATATAGTCGTCCGATCTGGCCCGTTCCGACATATATAGCA-GTGAGAGTATATA-GAAGACGGACAGACGG | |
| droFic1 | scf7180000449205:49213-49408 + | AT---------------------------------------------------------------------------------TTTTGCTATGGCAGCTGTATGATATAGTTTTCCGATTTGGACTATATTT------------------------------------GGCAAGTATATA---TAAAACTGGTCCAGGCGTACAATCTGTAAGTTTGGTTTACATA------------------------TCTTGAAAAACAAAAAAGTT----------TTTCATACTATATACT--ATTTTTATCCTATCGTTCCTATGGCAGCTATATGATATAGGCGTACGATGTGGC--------------------------------------------------- | |
| droEle1 | scf7180000491315:14317-14542 - | AT---------------------------------------------------------------------------------TGTTCCTATGGGAGCTATATGATATAATCGTCCGATTTTGATGAAATTTAAACCGTAATTCTGAAATATTTAACTAATCCTATATGTCTAAGAAG-------------------------------------------------------------------------TAAGATAAAAATTAAATTT----------T-TTTAT----------T-TATTTTTCCGATTGTTGCTATGGGAGCTATATGATATAGTCGTCCGATCCGGCTCGTTCCGACTTGTATACTA-CCTGCAATAGAAATAAAGATGGACGGAAAG | |
| droRho1 | scf7180000769384:24388-24586 - | AT-----------------------------------------------------------------------TTTG--ACATCGTTTCTATGACAGCTATATGATATAGTTGTTCGATTTTAATTTAATTAAAACCGTAATTCCATATTATCTAAAGATATCTATCTGTA-------------------------------------------------------------------------------------TA---------T----------TCT-TAAAT----------T-GTTTTTTGCGATTATTCCTATGGGAGCTATATGATATAGTCGCCCGATCCGGCT----------TATATACTA-CCTGCAATAGAAA----GACAACTTTTGAG | |
| droBia1 | scf7180000302049:53383-53636 + | CC------------------------------------------------------------------CTAATTTTG--TCATCGTTCCTATGGCAGCTATATGATATAGTCGTCCGATTTTGAAAAAATTTAATTTGAAATTCAGAACTAATTAAGAAATGTTATTTCTAAGCTTAGA-AG--------------------------------------GTAATA------------------------TGTTAAAAAGTACCGAAGATATAATTTTTTT-TAAGT------------TTTTCCTCCGATAGTTCCTATGGGAGCTATAAGATATAGTTGTCCGATCCGGCTGGTCCCGACTTGTACACTA-CCTGCAACAGAAA----GAAGGCCTTTGGA | |
| droTak1 | scf7180000415786:41288-41570 + | AATTTACAAATCGCAAAAATGTTAAT--TTTCCTATTA-----------------------TTTCTCATTAATTTTG--CGTTCGTTTCAATGGCAGCTATATGATATAGTAGTTCGATTTTGATAAA-------------------------------------TATCCTAGAGTAGA-AG--------------------------------------AAAATACAATAAAAACCAACGAATGTTATAGGTTCAAAAACACCCAAGATATAATTTTTTT-TACAT----------T-TTTTTTCCCGATTGTTCCTTTGAGAGCTATATGATATAGTTGTCCGATCCGGCTCGTTCCGACTTATATATTA-CCTGCAATAGAAA----GAAGACTTTTGGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/16/2015 at 08:48 PM