
ID:dvi_14660 |
Coordinate:scaffold_13042:3644197-3644347 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
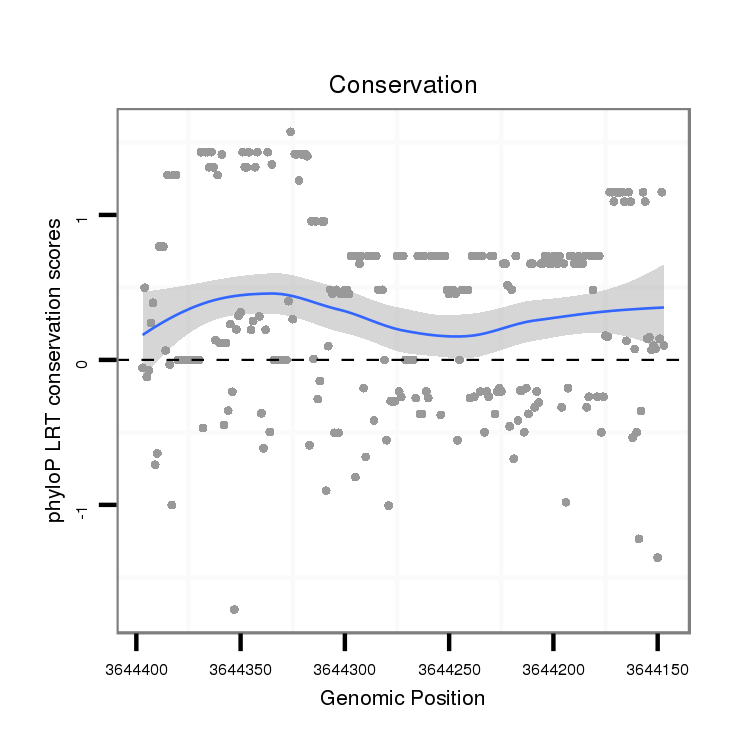
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ15602-cds]; exon [dvir_GLEANR_16047:1]; intron [Dvir\GJ15602-in]
No Repeatable elements found
| ------------------------------####################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATCCTTGAATACACGAAAGAAGTCAACAAAATGTCAAGACCCAATCAAAGGTTAGTGTAAAGAAAACCAAAAAAAAAAAAAAAATGATTCTCGTGCGAAGTTCAGTGCAAAAAAAAAAAATAACTAGAAAATAGGAATAAAAAAAAACATGTTATAAAAAATCTATAAACTGTGGAGGAATTTAGGCGGCCGCTAGCAACACCCGATGCAGCTGAAAAATAATAATCTTATCGACTTTGAACATTGTGTAC **************************************************(((((((...................................((((........))))...............(((.....)))...............(((..((((...((((((...))))))....)))).)))..)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
SRR060665 9_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
V047 embryo |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR1106722 embryo_10-12h |
SRR060679 140x9_testes_total |
SRR060658 140_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........ATACACGAAAGAAGTCAAC................................................................................................................................................................................................................................ | 19 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TGAATACACGAAAGAAGTC................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CGAAGATCAGTGCAAAAAAA........................................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AAGAAGTCAACAAAATGTCAAGACCC................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......AATACACGAAAGAAGTCAAC................................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AAGAAAGAAGTCAACAAAAT........................................................................................................................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CAACAAAATGTCAAGACCCA................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................AAAATGTCAAGACCCAATCAAAGC........................................................................................................................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................AATCTTATCGACTTTGAACATTGT.... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AACAAAATGTCAAGACCCC................................................................................................................................................................................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CCCGATGCACCTGAAA.................................. | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GAGAAGTTCAGTTCAACAAAAA....................................................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TAGTTGAGTGCAAAAAAAAAA..................................................................................................................................... | 21 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................AGAAGTCAACGAAATGTA........................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................AAAATAGGAAAAAAAAAAAA........................................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................AAAGAGGAATAAAAAAAA......................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAGGAACTTATGTGCTTTCTTCAGTTGTTTTACAGTTCTGGGTTAGTTTCCAATCACATTTCTTTTGGTTTTTTTTTTTTTTTTACTAAGAGCACGCTTCAAGTCACGTTTTTTTTTTTTATTGATCTTTTATCCTTATTTTTTTTTGTACAATATTTTTTAGATATTTGACACCTCCTTAAATCCGCCGGCGATCGTTGTGGGCTACGTCGACTTTTTATTATTAGAATAGCTGAAACTTGTAACACATG
**************************************************(((((((...................................((((........))))...............(((.....)))...............(((..((((...((((((...))))))....)))).)))..)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
GSM1528803 follicle cells |
SRR060670 9_testes_total |
M047 female body |
M027 male body |
V053 head |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................CTGGCGATCGGTGTGGACTA............................................ | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GCACGCTTCAGGTCACGA.............................................................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CTTTTTTTTTACTAGGAGCAG............................................................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CTGTCTTCGGTTGTTCTACA......................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| ....................................................................................................................................................................................................GTTGTGGGCACCGTCGAATT................................... | 20 | 3 | 15 | 0.13 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................GATCTTTTTTTTTTTTGACTAAGA................................................................................................................................................................ | 24 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGGTTATTTGATACCTCCT........................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:3644147-3644397 - | dvi_14660 | ATCCTTGA--ATACACGAAAGAAGTCAACAAAATGTCAAGACCCAATCAAAGGTTAGTGTAAA----------------------------------------------------------------GAAAACCAAAAAAA-------AAAAAAAAATGA------------TTCTC----GTGCGAAGT-TCAGTGCAAAAAAAAAAAATAACTAGAAAATAGGAATAAAAAAAA---------------------------------------------------------ACATGTTATAAAAAATCTATAAACTGTGGAGG------------AATTTAGGCGGCCGCTAGCAACACCCGATGCAGCTGAA----------AAATAATAATCTTATCGACTTTGA--ACA-TTGTGTAC |
| droMoj3 | scaffold_6308:867093-867364 - | ATCCTTAA----------------------AAATGTC------CATTCAGAGGTTAGTAAGAA---------------------------------------------------------------AAA-------AAAAA--------AAAACAAACAAAACAAATCAAGATGCTCAGCTATACGAAGTGTGAGTGAAAATAAAA-TCGCGATCA-----AAAAAAAAAAAAAAATAATAAT----------------------AAGGTACAAACAAAAATAATGAGAAATTACATGA-ATAATAGATTTCTGAATTGTGG----------------AATTCGACGTTCGCTAGCAACAACTGATGCAGCTTGACAAAATCAAGAAGTCGTAATCTTATCGACATGCGCCACATTCATCTAT | |
| droGri2 | scaffold_14853:2565176-2565443 - | ATCCTTGG-----AA-TAA-----------ATATGTCAAGAACAACCGTAAGGTTAGTAAGAAAG--------ATTAGAATGAACAAAAAGAAAAACACTTAACACCAAATCGAGTACGTGACCAAAAA-------CA---------------------------------------------------T-TGAGTAAAAATA-----TGCGACTA-----TAAAATTAAAAACAAAAACACT----------------------AAAATCCAAATTA--------------------------AAAATCTAAAAATAAAGGAAGAAGAAGCAGCAGCAACCAAACGGCAGCTAGCAACACCAAATGCAGCTGAA-----------AATAATAATCTTATCGACTTTAG--ACAGTTGTTTAC | |
| droBip1 | scf7180000395304:1861-1927 + | ATCCTTAG-----AATCAA-----------ATATGTCGAAAGTGTCGCAAAGGTTAGTAGAAA-------------------------------------------------------------------------AAGAA-------AAAAACAGACGA------------TTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453926:322672-322722 - | GTTAAT----------------------------------A----------------------------------------------------------------CCAAATAAAAATTTTTACCAAAAA-------AAAAAAAAAAAAAAAAAA-------------------AT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | x:7547901-7548048 - | ATTTAGCCAAATAAACGAA-----------ACATGTCGAAAGTGACACAAAGGTGGGTAATCA-CCGAAACAGAGTGC----------------------TACCA------C-CAGTTGGTAACAAATA-------AAAAAAAAAGGAAAAA-------------------------------------------------------------------------------------AATAATAAAAAAAAAAAAAAAACTTAAAAGAGTGTG------AATC------AA----------------------------------------------------------------------------------------------------CT------------------------------ | |
| droSec2 | scaffold_44:59533-59702 - | ATTTAGCCAAATAAACGAA-----------ACATGTCGAAAGTGACGCAAAGGTGGGTAATCAACCAAAACAGAGTGC----------------------TACCA------C-CAGTTGGTAACAAATA-------AAAAAAAAAAGAAAAAAA-----------------------------------------------------------------------------------------------AGAAAAAAAAATTAAGAGAGTGTGAGCATGAATC------AA----------------------------------------------------------------------------------------------------CTATCTTATCAACGG--A--ACT-CTGGAAAC | |
| droYak3 | v2_chr2h_random_006:13691-13693 + | TAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/16/2015 at 08:28 PM