
ID:dvi_14643 |
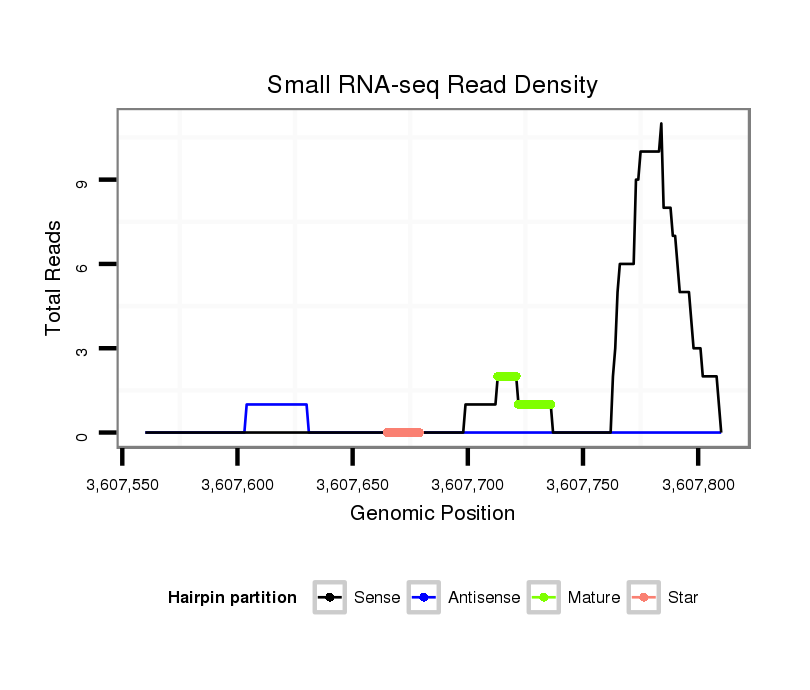
Coordinate:scaffold_13042:3607610-3607760 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
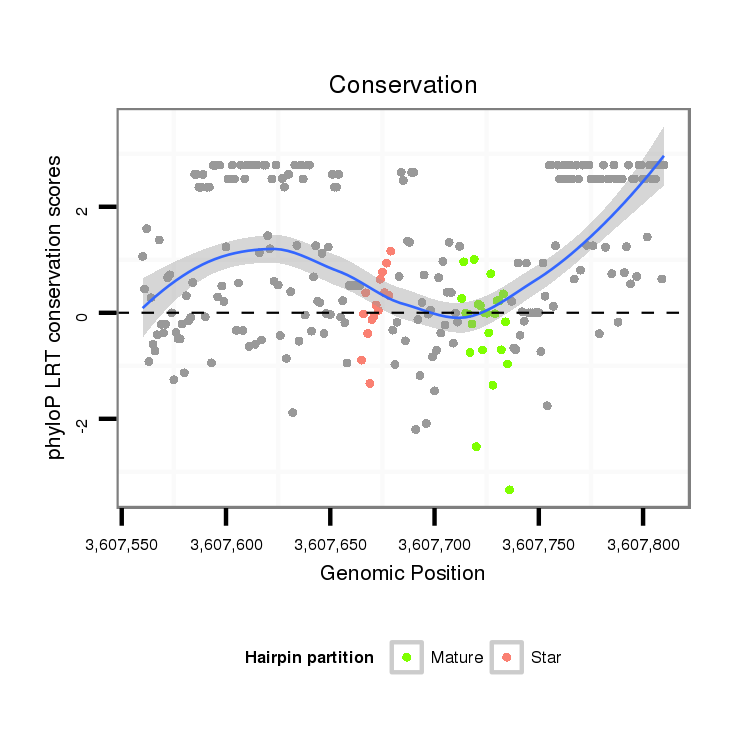
| -1.5 | -1.4 | -1.1 |
 |
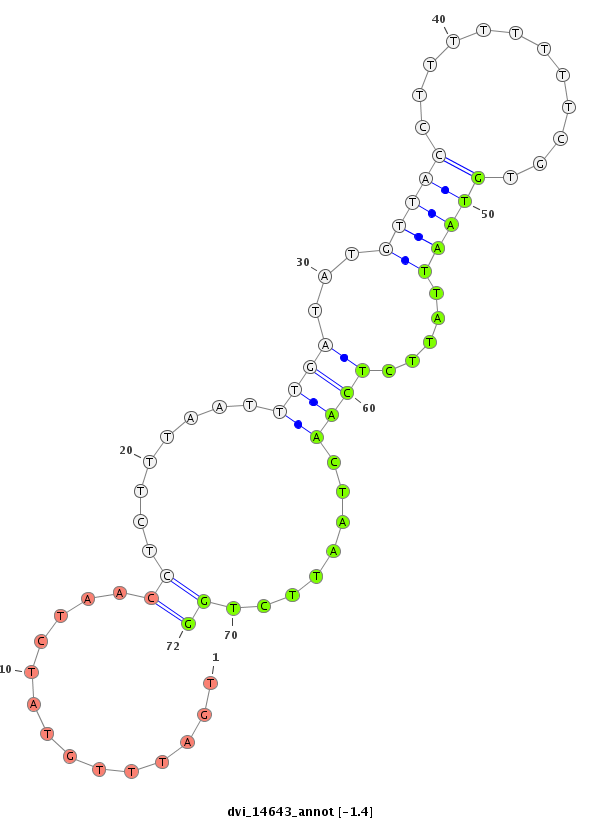 |
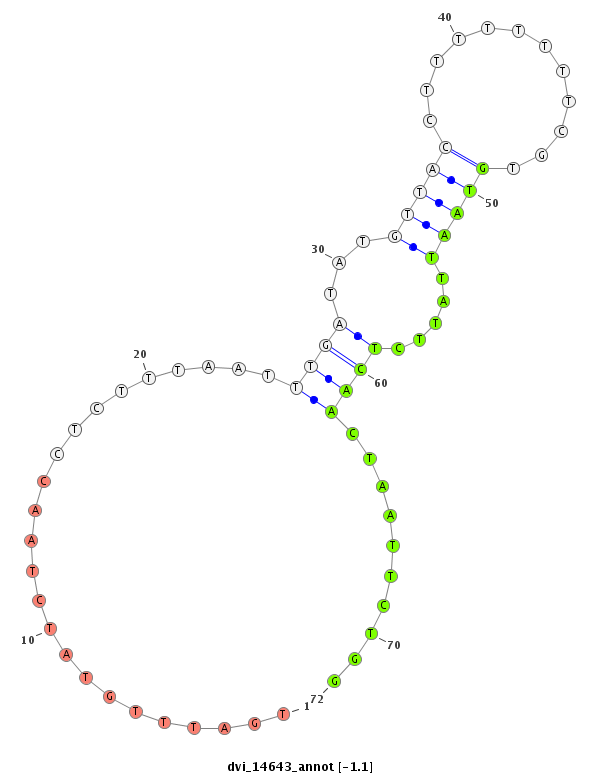 |
CDS [Dvir\GJ15970-cds]; exon [dvir_GLEANR_16396:3]; intron [Dvir\GJ15970-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAAAAGGGAAAACAAGCAAAGAAAAATGGATGCATTTACACGCACTTTTCATAAAACTTTTCGCACAACAACTACAACAAAAACCAGCCCAAGGTCTCAAATTTATGATTTGTATCTAACCTCTTTAATTTGATATGTTACCTTTTTTTTCGTGTAATTATTCTCAACTAATTCTGGTTTTCCCAAAAATTATGCTTGCAGCTCAACGACAAACTCGGCGCTGCCCTCACGCGTGCCCAAGCAACAGAATT *********************************************************************************************************......(((((...............))))).........................................************************************************************************** |
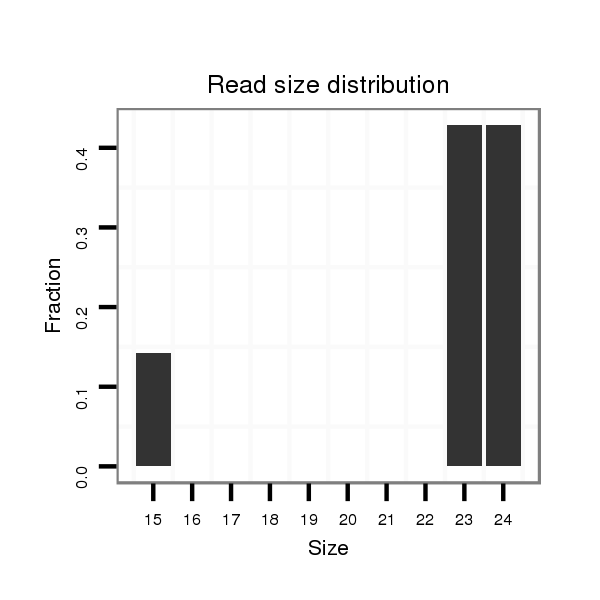
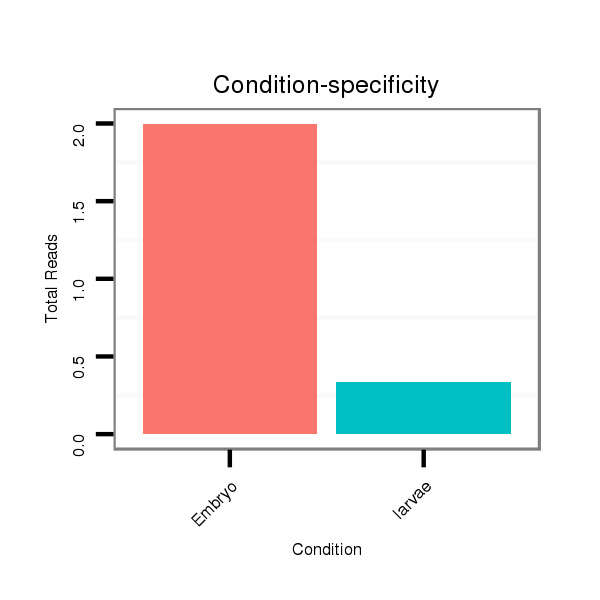
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M061 embryo |
M028 head |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060678 9x140_testes_total |
SRR1106729 mixed whole adult body |
SRR060662 9x160_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060666 160_males_carcasses_total |
SRR1106730 embryo_16-30h |
SRR060684 140x9_0-2h_embryos_total |
SRR1106727 larvae |
SRR060669 160x9_females_carcasses_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
SRR060685 9xArg_0-2h_embryos_total |
SRR060677 Argx9_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................CTCAACGAGAGACTCGGTG............................... | 19 | 3 | 20 | 11.35 | 227 | 90 | 2 | 17 | 20 | 13 | 0 | 0 | 3 | 28 | 22 | 4 | 0 | 0 | 0 | 15 | 0 | 0 | 5 | 0 | 3 | 2 | 2 | 1 |
| .........................................................................................................................................................................................................CTCAACGAGAGACTCGGCG............................... | 19 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................ACGACAAACTCGGCGCTGCC.......................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGAGAGACTCGGC................................ | 18 | 2 | 2 | 2.00 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGAGAGACTCGGGG............................... | 19 | 3 | 8 | 1.13 | 9 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CTCACGCGTGCCCAAGCAACAGAAT. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGACAGACTCGGTGA.............................. | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AACGACAAACTCGGCGCTGCCCTCACG.................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CTCGGCGCTGCCCTCACGCGTGCCCAAGC......... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CACCAGCTCAACGACAAACTCGGCGC.............................. | 26 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................CGGCGCTGCCCTCACGCGTGCC.............. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................GTAATTATTCTCAACTAATTCTGG.......................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CTCGGCGCTGCCCTCACGC................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CGACAAACTCGGCGCTGCC.......................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CAACGACAAACTCGGCGCTGCC.......................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TAATGCTGATTTTCCCAAAAA.............................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................CCTCACGCGTGCCCAAGCAACAGAA.. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CAACGACAAACTCGGCGCTGCCCTCA...................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CTACAACAAAAACCAGCTCAAGGTCT.......................................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CTCGGCGCTGCCCTCACGCGTGCCC............. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................ACCTTTTTTTTCGTGTAATTATT......................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGACAGACTCGGT................................ | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................TGATTTGTATCTAAC................................................................................................................................... | 15 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................ACCACAAAGACCAGCTCAAGG............................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......GGAATCAAGCAAAGAAAAAT................................................................................................................................................................................................................................ | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CAACGACTACAACAATAACCTGCC.................................................................................................................................................................. | 24 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGAGAGACTCGG................................. | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGAGATACTCGGTG............................... | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................TTTCAACTAAGTCTGGTT........................................................................ | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCAACGAAAGACTCGGGG............................... | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CCTATTTTTTCGTGTACT............................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTTTTCCCTTTTGTTCGTTTCTTTTTACCTACGTAAATGTGCGTGAAAAGTATTTTGAAAAGCGTGTTGTTGATGTTGTTTTTGGTCGGGTTCCAGAGTTTAAATACTAAACATAGATTGGAGAAATTAAACTATACAATGGAAAAAAAAGCACATTAATAAGAGTTGATTAAGACCAAAAGGGTTTTTAATACGAACGTCGAGTTGCTGTTTGAGCCGCGACGGGAGTGCGCACGGGTTCGTTGTCTTAA
**************************************************************************......(((((...............))))).........................................********************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060660 Argentina_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
V047 embryo |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................GAAAAGTATTTTGAAAAGCGTGTTGTT.................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................ACGGGAGTGCGCACGCGTTCGT........ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GTGCGCACGGCCTCGGTGT..... | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 3 | 0 | 1 |
| .....................................................................................TCGGATTCCAGAGATTAACT.................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:3607560-3607810 + | dvi_14643 | CAAAAGGGAAAA-CAAGCAAA---------------GAAAAATGGATGCATTTACACGCACTTTTCATAAAACTTTTCGCAC------------------------------------------------------AACAACTACAACAAAAACCAGCCCA---AGGTCTCAAATTTA---------------T--G-------AT---T-----T----------GTATCTAA-------------CCTC-TTTAATTTGATATGTT---------ACCTT------------------TTT----------TTTCGTG--------------TAATTAT------------------------------------T-------CTCAACT---------AATT----------C----------------------------T-------------GG---TTTTCCCAAAAATTATGCTTGCAGCTCAACGACAAACTCGGCGCTGCCCTCACGCGTGCC---CAAGCAACAGAATT |
| droMoj3 | scaffold_6308:832609-832864 + | CAAAGACAAAAAACA-CTCCA---------------TCGAAATGGAAGCCTTTACACGCACTTTTCACAAAACTTTTCGCACA-ACTT------------------------------------CCACA-----ACAACGAAAACCACAAAAACTAGCTCA---AGGTGTTT----------------------------------------------------------CTAACAATTGTGTATAACTCA-TT-CATTT-----ATTATTT-----TCTCA---------------TTGCAC----ACAAATTTTAACT--------------AAATTTTTATACTGAATTTGAA------------------------------------------------------------------------------------------------------ACATTT---------------TCTTGCAGCTCAACAACAAACTCGGCGCTGCCTTCCCGAGTGCC---CAAGCCACAGAACT | |
| droGri2 | scaffold_14853:10135525-10135809 + | CAATATCGATCTGCA-TTCCGATTTGAATTTGGTTGACGTAATGGAGGCATTTACACGCACCTTTCATAAAACGTTTCGCACG----AGCAACACA---------AC---------AA------CAACA-----TCAACAAGAACAACAAAAACTTCGACA---AGGTGTACAATTTATGATTTTGTAGAAATA--AATTATGTAC---G-----CAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTAATTAGT---------TATGTCACCGTAACCAATACGCTGTATTCATAT-TTGC-AT---TATTTA---------------TTTTGCAGCTCAACGACAAACTCAGCACTGCCATCGCGAGTACC---CAAGCAACAGAATT | |
| droWil2 | scf2_1100000004909:6555721-6556017 - | AA--------------------------------------TATGGAAGCCTTTAAGCGCACATTTCACAAAAGTTTTAGCACA-ACTAGAAATAGTAATAATATTAATACCACAACAACAACAACAA--CAAGAACAACCACAACAACAAATTCCTCAACA---AGGTCAAC---------------------CCAA-------AA---T-----C----------ATCCTTGC-------------CACA-ATTAAATTTTTTAG-------------TTGTTTTCCTTTTTTTTG---TAT----------T---ATT--------------TTGTTTC--AA-TAACTAGTCGAG------------------------------A-------------ATTATATTA----A----------------------------AGTTTCTTTATTCTG---AT---------------TTTTCCAGCTCAACAACAAACTCGGCGCTGCCATCACGAGTCCCATCCAAGCAACAGAACT | |
| dp5 | XL_group1a:5649891-5650101 + | ----------------------------------------------------TAGGCGCACATTCCAAAACACTTTCCGCACACACCAAAA---------------------------------------------------AACAACAAAATTCCCTCCT------------------------------------------------ACTCCCC----C------TC-----CCAGTGTGGATAATTAT-TTTGATTTTA------AACT-----ATTTG------------------TTT----------TCA--TT--------------TAATTTG----ATTGA--------TTTGCAACTTTTC-------------------------------------------------------------------------TAT-TGGC-TTAT-GTC-AT---------------TATTTTAGCTCAACGACAAACTCGGCGCTGCCATCGCGAATCCC---CAAGCAACAGAACT | |
| droPer2 | scaffold_63:297403-297615 - | --------------------------------------------------TTTAGGTGCACATTCCACAACACTTTCCGCACACACCAAAA---------------------------------------------------AACAACAAAATTCCCTCCT------------------------------------------------ACTCCCC----C------TC-----CCAGTGTGGATAATTAT-TTTGATTTTA------AACT-----ATTTG------------------TTT----------TCA--TT--------------TAATGTG----ATTGA--------TTTGCAACTTTTC-------------------------------------------------------------------------TAT-TGGC-TTAT-GTA-AT---------------TTTTTTAGCTCAACGACAAACTCGGCGCTGCCATCGCGAATCCC---CAAGCAACAGAACT | |
| droAna3 | scaffold_13417:1140894-1141160 + | AGA-----------------------------------GAAATGGAGGCTTTTACACGCACTTTGCAAAAAACGTTTCGCAGC-ACTAGGA------------------------------CCACGA--CCAGCACTACCACCACTACCAATTCGTCCACCACTAGGTGATC---------------------CCAC-------TA---T-----C----------CTC-----CCATTGTGTATAACAGTTTTTGATTTTGA-----GTC------ACTTT---------------------TGATATGATTTTAAGTCTCTTTTTTTTCGTATAACCTT------TAA--------CCTGTGAC--------------------------------------------------------------------------------------CTT---GAT-AT---------------CCTTACAGCTCAACGACAAATTCGGCCCTGCCATCGCGAATCCC---CAAGCAACAGAACT | |
| droBip1 | scf7180000395529:232793-233067 - | AG-------------------------------------AAATGGAGGCCTTTACACGCACTTTGCAAAAAACCTTTCGCAGC-ACCAGGA------------------------------CCACGA--CCAGCACCACCACAACCACCAATACCTCCACCACTAGGTGATC---------------------CCAC-------TACCTCCTCCCC----T------TC-----CCATTGTTTATAACTTT-TTTGATTTGAG------TC------ATTTG---------------------TGATATGATTTTA-GT----TTTTTATCATTTAATGT-----ATTAA--------CCTTTAACCTTTA----------------------A--------------------------------------------------AA------CTT---GCT-AT---------------CCTTGCAGCTCAACGACAAATTCGGCCCTGCCATCGCGAATCCC---CAAGCAACAGAACT | |
| droKik1 | scf7180000302696:444542-444814 + | AGAGAAGCA---------------------------ACAGAATGGAGGCCTTTACGCGCACACTACACAAGACCTTTCGCACT-ACTAAAACTACGACTAGTAGCAC---------GA------CCACC-----ACCACCACCATGACTAACTCCTCAAAT---AGGTTCTG---------------------C--T------------------T----------GCTCCCAA-------------CACT-TTTGATTTTCATTTTT------------TT---------------------CGACACTATCTCT-GT---------------------------------------TTTGATA--------TTTCTTTGAATACC----CAGTTCTAA--------------------------------------------CCA-GCTCCTT---GGTTTC---------------TCTTGCAGCTCAACGACAAACTCGGCCCTGCCATCCCGAATCCC---CAAGCAACAGAACT | |
| droFic1 | scf7180000454073:1785965-1786213 + | AG--------------------------------------AATGGAGGCTTTCACGCGCACACTGCAAAATACCTTTCGCAGC-ACAAGAA------------------------------CCACGACT-----ACAACTACCACCACAAAATCCTCCACC---AGGTTCCT---------------------CTAC-------AC---T-----G----------ATTCCTAA-------------CACT-TTTAATTTAAACTCTAATTT-----TTATG------------------TATTAATACGGTTTTA-GT----TTT--------------------------------TTTTCGGTTTTAA-------------------------------------------------------------------------CAC-TTTC-TTAA-CATCTT---------------TATTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droEle1 | scf7180000490751:1176115-1176355 + | AG--------------------------------------AATGGAGGCTTTCACGCGCACATTGCATAAAACCTTTCGTAGC-ACTAGAA------------------------------CCACGACT-----ACTACTACTACCACCAAATCCTCCACA---AGGTTCCC---------------------A--C-------AC---G-----C----------AACCCTAA-------------CACT-TTTGATTTAAATTTAAATCT-----TACTG------------------TTT----------T---------------------AATTTG----A--------TGACTTTTCAGTTTTAA-------------------------------------------------------------------------CAT-TGCC-AAAA-TTATTT---------------TGTTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droRho1 | scf7180000761121:132021-132247 - | AG--------------------------------------AATGGAGGCTTTCACTCGCACTCTACATAAAACCTTTCGTAGC-ACTAGAA------------------------------CCACGACT-----ACTACCACTACCACCAAATCCTCTACC---AGGTTCCT---------------------C--C-------AC---G-----A----------ATCCCTAA-------------CACT-TTTGATTTCTTTTTAA-TTT-----GACCG------------------TTT----------TTCAGT-----------------------------------------------TTTAACATTTTT-------------------------------------------------------------------------C-ATAA-TTATTT---------------TATTGCAGCTCAACGACGAACTCGGCTCTCCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droBia1 | scf7180000302069:1101473-1101716 - | AG--------------------------------------AATGGAGGCCTTCACGCGCACTCTGCAAAATACCTTTCGTAGC-ACAAGAA------------------------------CCACGA-----------CCACTACTACCAACTCCTCCACC---AGGTTCCT---------------------C--C-------AC---A-----C----------ATCCCTAA-------------CCAC-TTTGATTTAAC--------CATTGGAATTG------------------TTT----------T---ATT--------------TTGGTTC--AGCTCCCTTGTGTAC------------------------------TCCTAGTTT-------------C----A----------------A-----------CGT-TCTCCAC---CCTTCC---------------TTTTGCAGCTCAACGACGAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droTak1 | scf7180000415169:792863-793123 - | AG--------------------------------------AATGGAGGCCTTCACGCGCACTCTGCAAAATACTTTTCGTAGC-ACCAAGAGAACC---------AA---------AA---CCACGA--CTACTACTACCACCACCACCAACTCCTCCACC---AGGTTCCG---------------------A--C----------------------------------CAG-------------CACT-CTTGATTTCACTTATAATCT-----TTCTG------------------TTT----------TTTGGTT--------------TAAGTTTTAAGTTTGTTTGGGAACTTTTTAATTTCAA-------------------------------------------------------------------------CAT-TATC-CC---GACTTT---------------TCTTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droEug1 | scf7180000409230:828867-829133 - | AGACAAGCA---------------------------TCAGAATGGAGGCTTTTACGCGCACACTTCAAAAAACGTTTCGTAGC-ACTAGAA------------------------------CCACGA--CTACCACTACTACTACCACCAATTCCACCTCC---AGGTTCCT---------------------C-AC-------CA---G-----C----------ATCCCTAA-------------CAGT-TTTAATTTCAATTCGTATCT-----TTGTA------------------TAT----------GTTATTT--------------TCGGTTC------CATTTGAGTATTTCTTTGTTTTAA-------------------------------------------------------------------------CCT-TAGT-TT---TTAATT---------------TTTTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| dm3 | chrX:19504196-19504463 + | -----------------------------------------ATGGAGGCCTTTACGCGCACACTGCAAAATACCTTTCGCAGC-ACTAGGACCACC---------AG---------AA------CCACT-----ACTACAACAACCACAAACTCCTCCACC---AGGTTCCT---------------------A--CCTCACCTAC---G-----C----------AACCCTAA-------------CACC-TTTGGTTTTATTTCGAGTCT-----ACTTG------------------TTC----------TTAAGTT--------------TCGCTTC----------------------------AA------------CACT----CAACTGCAATACTCATGTACAAT-A----------------------------TGTT----------------CGTCTCA----ACTTATTTGCAGCTCAACGACAAACTCGGCGCTGCCATCCCGAATCCC---AAAGCAACAGAACT | |
| droSim2 | x:18397615-18397877 + | dsi_4328 | -----------------------------------------ATGGAGGCCTTCACGCGCACACTGCAAAATACCTTTCGCAGC-ACTAGGACCACC---------AG---------AA------CCACT-----ACCACCACTACCACAAACTCCTCCACC---AGGTTCCT---------------------A--CCTCACCTAC---G-----CAACCCTAACA---CCTAA-------------CACC-TTTGGTTTTATTTCGAGTCT-----ACTTG------------------TTC----------TTAAGTT--------------TCGCCTT------------------------------------A-------CT----TAGCTGCAATACTT----------A----------------------------TGTT----------------CGTCTTA----ACCTATTTGCAGCTCAACGACAAACTCGGCACTGCCATCCCGTATCCC---AAAGCAACAGAACT |
| droSec2 | scaffold_8:1784907-1785169 + | -----------------------------------------ATGGAGGCCTTCACGCGCACACTGCAAAATACCTTTCGCAGC-ACTAGGACCACC---------AG---------AA------CCACT-----ACCACCACTACCACAAACTCCTCCACC---AGGTTCCT---------------------A--CCTCACCTAC---G-----CAATCCTAACA---CCTAA-------------CACC-TTTGGTTTTATTTCGAGTCT-----ACTTG------------------TTC----------TTAAGTT--------------TCGCCTT------------------------------------A-------CT----TAACTGCAATACTT----------A----------------------------TGTT----------------CGTCTTA----ACCTATTTGCAGCTCAACGACAAACTCGGCACTGCCATCCCGTATCCC---AAAGCAACAGAACT | |
| droYak3 | X:11832626-11832880 - | AG-----------------------------------CAGCATGGAGGCCTTCACGCGCACACTGCAAAACACCTTTCGTAGC-ACTAGGACCACC---------AG---------AA------CCACT-----ACTACCACAACCACAAACTCCTCCACC---AGGTTCCT---------------------T--C-------AC---G-----C----------ATCCCTAA-------------CACT-TTTGATTTTATTTGGAATCT-----ACTTG------------------TTT----------TTAAGTT--------------TCGTTTC------------------------------------A-------CT----CAATTGTAATACTT----------A----------------------------TGTG----------------CATTTCA----ACCTATTTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---CAAGCAACAGAACT | |
| droEre2 | scaffold_4690:9732397-9732649 + | AA-------------------------------------GCATGGAAGCCTTCACGCGCACACTGCAAAATACCTTTCGTAGC-ACTAGGACCACT---------AG---------AA------CCACT-----ACTACCACTACCACAAACTCCTCCACC---AGGTTCCT---------------------T--C-------AC---G-----C----------ATCCCTAA-------------CACT-TTTTATTTTATTTGGAAACT-----ATTTG------------------TTT----------TTAAGTT--------------TCGCTTT------------------------------------A-------TG----TAATTGTTATACTC----------A----------------------------TATT----------------AATTTCA----ACCTATTTGCAGCTCAACGACAAACTCGGCTCTGCCATCCCGAATCCC---AAAGCAACAGAACT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:25 PM