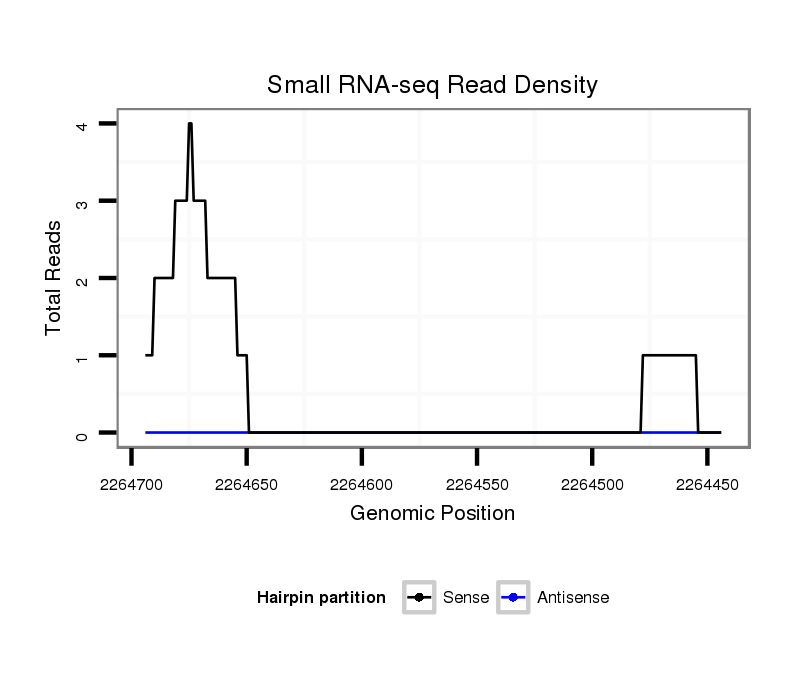
ID:dvi_14400 |
Coordinate:scaffold_13042:2264494-2264644 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_16119:3]; CDS [Dvir\GJ15679-cds]; intron [Dvir\GJ15679-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACCCATATCCAGTGTGCTGGAGGAGCTGCTCAAAAACACCGGCTATGATGGTAAGTAAGCAATTCCTTAAACCTAAGTTGGAATTGTTGGGAGCATGCGGCTTGACAGGCAGCTCACTGATTAGTTGCGTTCAAGGATTTTCCTGGCATCAACAATGCGGTTTAATAGTGTGAAAGATCCTGGAAGATGGAAAATCAATTCGGCAATTTGTTGGCCATTCTCGCTGAACTGCTTTCCCCATGTTTCTTGTA **************************************************.......(((((((((.............)))))))))..((((..((((((...(((........)))...))))))))))....(((((((.((.(((....((((.........))))....))))).))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
GSM1528803 follicle cells |
M047 female body |
SRR060658 140_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060676 9xArg_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060678 9x140_testes_total |
SRR060683 160_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................AATGCGGTGTAATCGCGTGAA............................................................................. | 21 | 3 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................GAGGAGCTGCTCAAAAACACCGGCTA.............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AATTCTCGCGGAAGTGCTTTCCCC............ | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ATATCCAGTGTGCTGGAGGAGCT................................................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................AACACCGGCTATGATGAT....................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GTGCTGGAGGAGCTGCTCAAAAACACC................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAATTGTTGGGAGCATCTGGCT..................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ACCCATATCCAGTGTGCTGGA...................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................ATTCTCGCTGAACTGCTTTCCCCA........... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAATTGTTGGGACCATGTGG....................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AATGCGGTGTAATCGCGTGA.............................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................ATACACCGGCTAAGATTGT....................................................................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................AATGCGGTGTAATCGCGTG............................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................TGCTGGAAGATGGAAAAA........................................................ | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................CAATGCGGTGTAATCGCG................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AAACCTTAGTTGGATTGGT.................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGGGTATAGGTCACACGACCTCCTCGACGAGTTTTTGTGGCCGATACTACCATTCATTCGTTAAGGAATTTGGATTCAACCTTAACAACCCTCGTACGCCGAACTGTCCGTCGAGTGACTAATCAACGCAAGTTCCTAAAAGGACCGTAGTTGTTACGCCAAATTATCACACTTTCTAGGACCTTCTACCTTTTAGTTAAGCCGTTAAACAACCGGTAAGAGCGACTTGACGAAAGGGGTACAAAGAACAT
**************************************************.......(((((((((.............)))))))))..((((..((((((...(((........)))...))))))))))....(((((((.((.(((....((((.........))))....))))).))))))).............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
V053 head |
V047 embryo |
|---|---|---|---|---|---|---|---|---|
| ...........................CGAGTTTTTGTGCCCGAT.............................................................................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................CGCCCAGTGACTAATCTAC............................................................................................................................ | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ..............................GTTCTTGTGGCTGAGACTAC......................................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................................ATCTTCCACCTTTTACTTAA................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:2264444-2264694 - | dvi_14400 | ACCCAT---ATCCAGT-----GTG------CTGG-----------AGGAGCTGCTCAAAAACACCGGCTATGATGGTAAGTAAGC-------AATTCCTTAAACCTAAGTTGGAATTGTTGGGAGCATGCGGCTTGACAGGCAGCTCACTGATTAGTTGCGTTCAAGGATTTTCCTGGCATCAACAATGCGGTTTAATA------GTGTGAAAGATCCTGGAAGATGG-------------------------------------------------AAAATCAATTCGGCAA---------------TTTGTTGGCCATTCTCGCTGAACT--------------------------------------------GCTTTCCCCATGTTTCTTGTA |
| droMoj3 | scaffold_6328:1296605-1296673 + | GCCCAT---TAGTACA-----GTG------CTAG-----------ACGATCTGCTCAAGACAACTGGCTTTGATGGTAAGTAACA-----GAAATTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT | |
| droGri2 | scaffold_15203:6223809-6223811 + | GTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:1184127-1184189 + | TTCTGCAGCTTCCAAA-----GTG------ATTA-----------CCGAACTTCTCCAGTCTACAGGACATGCTGGTAAGTCAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13045:909687-909785 + | AATAGT---------------------------------------AGGGCTTATTAGTGC-----------------------------------------------------------------------------------------------------------------------------------------------CGATT---------TTAGATGTTC--------------------------------------------------GAAGGTAGTTTGACAG---------------TTAGCTGTCCACTCGAATTTTACA------------------------------------------TTTTTATACCCTTGCAGAGGGTA | |
| droBip1 | scf7180000395673:7016-7124 + | TTCTTTGC----AAC--------A------GAGT-----------TTGGTTTATTAATGC-----------------------------------------------------------------------------------------------------------------------------------------------TGATT---------TAAAATGTTC--------------------------------------------------AAAGGCGTTTTGACAT---------------TCTGTAGTCCATTCGAATTTTACA------------------------------------------TTTTTATACCCTTGCAGAGGGTA | |
| droKik1 | scf7180000298918:1234-1373 - | TCCTTCAGCCAGAAAA-----AGT---------------------ATTATTTTATA-TTT-----------------------------------------------------------------------------------------------------------------------------------------------TGAAA---------TCAAATTTTG--------------------------------------------------GAGGACAATTCGGAAA---------------GCTGTT-----------CTGAATTGGGTTTTCTACGCTTAACAAATCTGCATACTACATTTAGGGCTTTTTATACCCTTGCA--GGGTA | |
| droFic1 | scf7180000454072:751121-751159 - | GGA----------------------------------------------GCTGCTCGGACCCACGGGATACAGTGGTAAGTAAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000491033:157823-158048 - | ATATCA----AGTAAG-----CTG-----AACAG-----------AGGGTTTATTTAAAAAAACAAGAAAGAAAGCAAACAAAGCAAGCAAGCCG-----------AAGTTC------ATATACCCTTGCAGCT--ACAAGTATTGCAATAATTAAACATTTTTGAA---------AACATTAAAATTATGATTTACTTGCGTATATGTGAAACAACAT------------------------------------------------------------TGTAGCTATGATGATTTGCAGCTCAATAATTAGATA----------------G------------------------------------------TTATTATACCCTTCCAGAGGGTA | |
| droRho1 | scf7180000765085:458-627 - | CGGCTTGC----CGAAGTTAGCTTCCGTCCTCGTTGCGTTTATTTTTGATCTGTCG---ACAATCA---AGTTTGAAAACGAAACATTCAGGTCA-----------AAAT--------------------------------------------------------------------------------------------------------------------GTATAATTTCGCCGGTTTTTTTGTAAGTTGAGTATAGATCGCCAATTTTTGGAG------------------------------------------------AACA------------------------------------------TTTTTATACCCTTGCAGAGGGTA | |
| droBia1 | scf7180000301398:6534-6536 + | GTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000410408:25722-25817 + | TTTAGTAATGTGA----------------------------------------------AAAATTAAAGTGAATGAAAAAGAAAATATCAGATCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTTTGCATTCTTTAAATTTGTA------------------------------------------TTTTTATACCCTTGTAGAGGGTA | |
| droEug1 | scf7180000409522:331602-331674 + | TCCTT----ATCTGTT-----GCG------ATAA-----------CTGATCTGCTTAGACCCACCGGATACAGTGGTGAGTGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------------------------------AACCTATACCT----------TA | |
| droYak3 | X:2623807-2623854 + | TGT--------------------G------ATCT-----------TGGGTCTGTTAAAACCCACGGGATACAGTGGTGAGTGAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 04:18 AM