| droVir3 |
scaffold_13042:1670496-1670877 + |
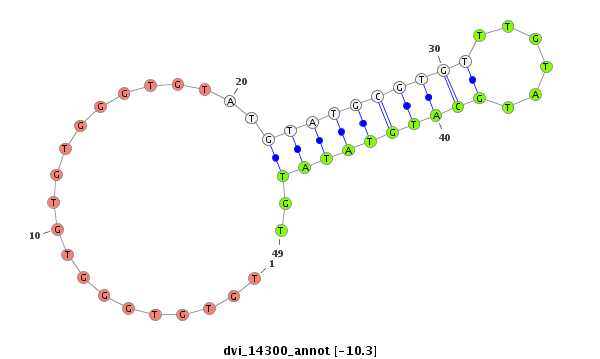
dvi_14300 |
GAG---------------------------------------GAGCGCCGGCACATACAGAAAGTGCAGAATGCCTTTCAGTACTACAGGTGG--------GTG-------------------C-------A--------------------------TGG--------------------------------AATTGTAAACAAACGGTATTTAAGC-----------------CAAACGCGCTG------ATGCAAATTA--CTACCCGCAGCAC---ACACTAACACA---------------CATATATGCACATATGCGTGCACATGTGT-------GTAT-------------------------------------------------------------------------------GCGTGCATGTGCATGTTCAACAGCTGATAACGAAAATCAATTTAAGCGGCTTAAC------------------ATGTGCACATGT--GTGCC------------------------------------------------------TGTGTGTCTGTGTGG--------G-----TGT-------------GTGGGTGTATGTATGCGTG-----------------TTTGTATGCA----TGTA---------------TATGTAGGACACTTGCT-----------------------------------CT----------------------------A-------------AT------------------CGTTTATTTTGCTCCAA-------------ACAG-------------------------A-------------------------------------CGTTATGCCTACCTAAG---------------GGTTAACAAAACGGAGGATTACCTTAA-CAGTT----------- |
| droMoj3 |
scaffold_6359:1602609-1602974 + |
|
GAG---------------------------------------GAACGGCGCCACATACAGAAAGTGCAGAATGCCTTTCAGTACTACAGGTGG--------GTC--------------------------------------------------TTCATGC--------------------------------AATTGTAAACAAACGGTATAAATGC-----------------CCAAAGTGCCA------ATGCAAATTA--C---C------------CGCTCACACA---------------CACATGTACACACA-----------------------------------------------------------------------------------CTCAGGGCTTGGGTAGAAGTGTGCATGAATGTGCATGTTCAACAGCTGATAACGAAAATCAATTTAAGCGGTTTAAC------------------ATGTGCCAACG------------T--------------------------AC------------------TTGTG----------TGA--------G-----TGT-------------GTGTGTGTGTGCTTAC-----------------------------A----TGTG---------------TATGTAGAACATCTGCT-----------------------------------CT----------------------------A-------------AT------------------CATTGACTTTATTCCCACACACATCACAAACTAG-------------------------A-------------------------------------CGTTATGCCTACATAAG---------------GGTTAACAAAACGGAAGCTTACCTCAA-CAGCT----------- |
| droGri2 |
scaffold_15074:3493730-3493948 + |
|
CAG---------------------------------------TATA---------------------------------------------------------------------------------CAGG--GTATACGCGTAAAAACA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCG---------CTAGCAG------------CACACAC--TCGTATATAAAT--ACTGACGAA--------CATT-TACCATCATGTGGC-----------------------------------------------------------TAATGCCT------------------CCATGTGCCTGTGTGTGTC----TGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG--TGTGT------GTG------------------TGTGTGTATATGTGTTGTTGTT-------------------------GT--------------------TG-TGTTTGCTTCAT-----------TCAG-------------------------------------------------------------------------------------------------------------------------------------TCTGTG--------------------------------------------------------------------------------------------------------------------------T--------GC |
| droWil2 |
scf2_1100000004585:7895650-7895885 + |
|
TGT---------------------------------------ATGC---------------------------------------------------------------------------------GCAT--GTATTTAAGTG----------------------------------------------------------------GCGTCAGCGG------------------------------------------------------------------------------------CAGACAGCTGAATGTACA-----------------------------------------------------------------------------------TTTAGCGTGC-----------------------------------------------------------TGTTTGCC------------------AGTTGTTTGTGCGTGAGTT----T--------------------------GA--------------------------CTCTGT--GTGTGTGTGTG-----TGT-------------GTGTG--------TGTGTG--CTTGTGT-----G--TTTGT----T----TGCC---------------AATGTATTACACTCGCC-----------------------------------AG----------------------------G-------------CTTGCCACTCCATCTCCATCTCTTTATCTTGCTC-----------------CAG-------------------------G-------------------------------------CGTAATGCCAACAGGAGCAGAAAGCAGCAAGAGCTTAA-------------------------------------C |
| dp5 |
XL_group1a:2482591-2483038 + |
|
GAA---------------------------------------GAGCA---GCACATTCAGAAGGTCCAGAATGCCTTTCTGTACTATGGGTGA--------GTG-------------------T-------T--------------------------TGCCTTTATGGCTGTCAGTGAAATGGTCTCCGGCCAACTGTATTTGAATGCTGATCGGGAGAGGTTGAGTCGGGGTGAAGAAGCGCTGTGACATGTGCAGACAAGTCCGCCGGCAGCTCAACAAATTACCGTAGTTTTACGC-------------------------------------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGGGTGCATTTTTGAGCGTTCGTGCGTTCGTG----------------CGT--------G-----CGT-------------GTGTGTGTGTACATGATAA-----------------GCAGC----CTG--------CA-TGTCTGTTCTTG-----------TACGGGACATGCAGTTGTCGCAACATGATTCGTACCACCAAACACACGGCGTCAATGACTGATAATGAAAATCAATTAATTTATCTGCCACT-CATCCGCACCCCGTATCC----------------------ACAG-------------------------A-------------------------------------CCATATGCATGCCAGCG---------------GCTGAGAAGAACCCTGGATTACCTCAA-CAGCC----------- |
| droPer2 |
scaffold_45:609937-610187 + |
|
CAC---------------------------------------ACAC---------------------------------------------------------------------------------ACAC--ACATAGGTGCATATGTAGGTG---------------------------------------------------------TGTA------------------------------------------------------------------------------------TGGTA--------G--GCGTAT----G-GCCCGTAT-------ATTT-----------------------------------------------------------------------------------------GTATGTTTT----CTGACCATTATACATGATTTCCTCGCTTTTGTAGGCCTCTCCTTCACTACTTACTCATCAG------------------------TGTGTGT--------GT------------------------GTGTGTGT--------GT---GTA--TGT-------------GCC------TG--TGCATG--CGTGTGT-----G--TGTGT----TTATTG---------------------GTAGAATGTTTGTT-----------------------------------GC----------------------------A-------------TT------------------TATGGGTACTGCTCCTA-------------GCAG-------------------------T-------------------------------------TGTTGTCCT-CCTGGAG---------------GAG-------------------------CAGCA----------- |
| droAna3 |
scaffold_13250:2996437-2996596 - |
|
TGA-TGTCATGCGCAACCGGCCGCACCTTG-------------CAC-----------------------------------------------A------------------------------TGTATATGTAT--ATGTATATGTATATGTA-------------------------------------------------------------------------------------------------------------------------------------------------TTGTA--------T-----------------------------A---TGTATATGTATATGTA---------------------------------------------------------------------------------------------------------------------T------------------GTATAT----------------------------GTATATGTATATGTATATGTATATGTATA---------------------------------------TG---------------TATATGTATATGTATATG-----------------TATATGTATA----TGTA---------------TA--------------------------------------------------------------------------------------------------------------------------------------------------------TG----------------------------------------------------------------------------------------------------------------------------TA------- |
| droBip1 |
scf7180000396676:36772-36904 + |
|
AC-------------------------------------------------------------------------------------------T------------------------------AACACAT--AC--ATACAT-------------------------------------------------------------------------------------------------------------------------------------------------------------TGTA--------T-----------------------------G---TATGTAAATGTATGTGTGTGTGTGTTAAATAATCGTAGAAAATCCTAG-----------------------------------------------------------------------------------CAT------------------GT------------GCGCTTGTAT--------------------------GT------------------CTGTG----------TGT--------G-----TGT-------------GTGTGTGTGTGTTGGAGTG---------------------------------------------------------------TCAG-------------------------------------------------------------------------------------------------------------------------------------TGTGAG--------------------------------------------------------------------------------------------------------------------------T--------GT |
| droKik1 |
scf7180000298888:10291-10450 + |
|
GT-------------------------------------------------------------------------------------------ATATATGTATA-------------------T-------ATGTATATATGTATATA--------------------------------------------------------------------------------------------------------------------------------------------------------------TGTATATATGTATATATGTATATATGTAT-------ATAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATATATGTATATATGTATATATGTATATATGTATA--TATGT------ATA------------------TATGTATATATGTATATATGTA-------------------------TA--------------------TA-TGTATATTTGTA-----------TATA-------------------------------------------------------------------------------------------------------------------------------------TGTATA--------------------------------------------------------------------------------------------------------------------------T--------AT |
| droFic1 |
scf7180000454066:3054238-3054382 + |
|
AACAC---TTGCGA-AATGAATTAACTTTAAGCCGCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---------------------------------------------------------------AATTT-----------------------GTGCTCTTGTGTGCGCT----CATATCTGTATGTGTGGATGA------------------------------------------GT--------G-----TAT-------------GTGTGTGGGTGCTCGTACGCTTGTGTGAGTGTG-CA--------------------TA-CATAAATGCTCG-----------TAAT-------------------------------------------------------------------------------------------------------------------------------------TGTTTA--------------------------------------------------------------------------------------------------------------------------T--------TT |
| droEle1 |
scf7180000490546:1027102-1027310 - |
|
GTAAATTAATGTGC-GCTGGATAAACTTTGTGTCGTCCATGTGTGT--------G---------------------------------TGTGTGTGTGTGTGTG-------------------T-------GTGTGTGTGTGTGTGTG--------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGTGTGTGTGTGTGTGTGTGTGTGT-------GTGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG--TGTGT------GTG------------------TGTGTGTGTGTGTGTGTGTGTG-------------------------TG--------------------TG-TGTGTGTGTGTG-----------TGTG-------------------------------------------------------------------------------------------------------------------------------------TGTGTG--------------------------------------------------------------------------------------------------------------------------T--------GT |
| droRho1 |
scf7180000779872:341211-341379 - |
|
CC-------------------------------------------------------------------------------------------GCAGGATTGCGTGACCAACACTTATGTGTA-T--GTAC--GTATTTGCG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTTGATGTGTGTGTGTGTGTGT-------GTGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT--------GTGTGTGTGTG------------------TG------------TGTGTGTGTGTGTGTGTG----TG---------TG----TGTGTG-----------------TGTG--CG------------------------------------------------------------------------------------------------------------------------------------------------------TGTG--------------------------------------------------------------------------------------------------------------------------GGTACTCTGAT |
| droBia1 |
scf7180000301232:32917-33075 - |
|
AT-------------------------------------------------------------------------------------------A------------------------------TGTATAT--GT--ATATGTATGTGTATATGTA------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTATATGTATATGTATGTGTATGTGT-------ATAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATATGTATATGTATATGTATATGTATATGTATGTG--TATATGTATGT--GT----------------------ATATGTATATGTATATG----TA-----------------------------------------TG-TGTATATGTATG-----------TGTA-------------------------------------------------------------------------------------------------------------------------------------TATGTA----------------------------------------------------------------------------------------------------------------------------TA------- |
| droTak1 |
scf7180000415381:115351-115505 - |
|
TAT---------------------------------------GTGT---------------------------------------------------------------------------------GTGT--GTGTGTGTGTGTGTGTGTGTG--------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGTGTGTGTGTGTGTGTGT-------GTGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG--TGTGT------GTG------------------TGTGTGTGTGTGTGTGTGTGTG-------------------------TG--------------------TG-TGTGTGTGTGTG-----------TGTG-------------------------------------------------------------------------------------------------------------------------------------TGTGTG--------------------------------------------------------------------------------------------------------------------------T--------GT |
| droEug1 |
scf7180000409509:172055-172230 + |
|
CAC---------------------------------------ACAC---------------------------------------------------------------------------------ACAC--ACACACATCTACACATACGCA--------------------------------------------------------------------------------------------------------------------------------------------------------------C----------------------------------------------------------CGAA--CTCGTCGAAAATTTCAAATTAAAATTCTGCAA---------------------------------------------------------------AGTTTGCC------------------ATGTTCTCGTG------------T--------------------------GT------------------GTGTGTGTGTGTGTGTGT--------G-----TGT-------------GCTAGTCTATGTGTGTGTG-----------------TTTGTGTTTG----GGCAAACACTGCGCATAATTT-----------TGCT-------------------------------------------------------------------------------------------------------------------------------------CAAATC--------------------------------------------------------------------------------------------------------------------------T--------TT |
| dm3 |
chrUextra:22197359-22197549 - |
|
CAAAT---CGATAC-C-----------TGTA------------ATTATTAACTTA---------------------------------TATGTATATATGTATA-------------------T-------ATGTATATATGTATATA--------------------------------------------------------------------------------------------------------------------------------------------------------------TGTATATATGTATATATGTATATATGTAT-------ATAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATATATGTATATATGTATATATGTATATATGTATA--TATGT------ATA------------------TATGTATATATGTATATATGTA-------------------------TA--------------------TA-TGTATATATGTA-----------TATA-------------------------------------------------------------------------------------------------------------------------------------TGTATA--------------------------------------------------------------------------------------------------------------------------T--------AT |
| droSim2 |
x:9559844-9559934 + |
|
GT-------------------------------------------------------------------------------------------G------------------------------TGTGTGT--GT--GTGTGTGTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGT--------GT------------------------GT--------GTGTGTGT---GTG--TGT-------------GTG------TG--TGCGAG--TATGTGT-----G--TGTGT---------------------------------------TTTGTG-----------------------------------CT----------------------------A-------------TT------------------CACGAATTTT------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_8:2731273-2731452 - |
|
AT-------------------------------------------------------------------------------------------A------------------------------TATAAGT--AT--GTGCGGGCATATGTATATT------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTATTTGTATATGAACG---------GCAACAG------------CGCACAC--AATTA--ATAAG--AATTAC-AA--------GTTG-TATGATGAGGGGGC-----------------------------------------------------------GGTTGGCT------------------GTGTGTCTGCGTG------------------------------------------------------------TGTGTGGGTGTGTGG--------G-----TGT-------------GTGGGTGTGTGTCTGTGTG-----------------TTTGTGCGGG----CTTA---------------TA--------------------------------------------------------------------------------------------------------------------------------------------------------CG----------------------------------------------------------------------------------------------------------------------------CA------- |
| droYak3 |
X:6918662-6918820 - |
|
CAT---------------------------------------ATTC---------------------------------------------------------------------------------TTAT--ATATATGCGTATACATATATA--------------------------------------------------------------------------------------------------------------------------------------------------------------TATAGCAA----------------------------------TATATGTGTTTGTGTGGAAA----------------------------------------------------------------------------------AAAAAATTATTCGTGCGGTTTTCT------------------CTATGCATACGTGTGTGCC------------------------------------------------------T--GTGTGTGT--GT---------GTACGTGT-------------CTGTGTGTG----TGTGGCTCTATG--T-----G------GGTGTG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGCG----------------------------------------------------------------------------------------------------------------------------TG------- |
| droEre2 |
scaffold_4770:5918676-5918930 - |
|
AT-------------------------------------------------------------------------------------------G------------------------------TATGTAT--GT--ATGTATGTATGTATGTATG------------------------------------------------------------------------------------------------------------------------------------------------------TATGTATGTATGTATGTA------TGTAT-------GTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATGTATGTATGTATGTATGTATGCATGTATGTA----T--GTATGT--ATGT------------------ATGCATGT----ATG---------------------------------TATGT----ATGTACA----TA-TGTATGTA--TG-----------TAC--------------------------------------------------------------------------------------------------------------------------------------TGTATGTGTTACTTGCACAAAGTAGTAAAAATTGTCTTAATTAAAGCCAAATAACCGGGCAAATGTTATGCATACAAGTG---------------TTTGAAATGTTATAATATTTATTTTAAAATATT----------- |