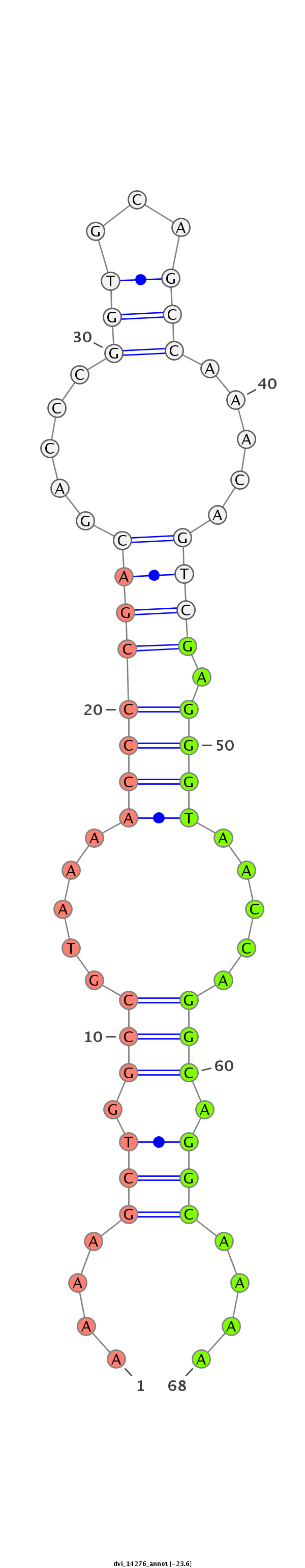
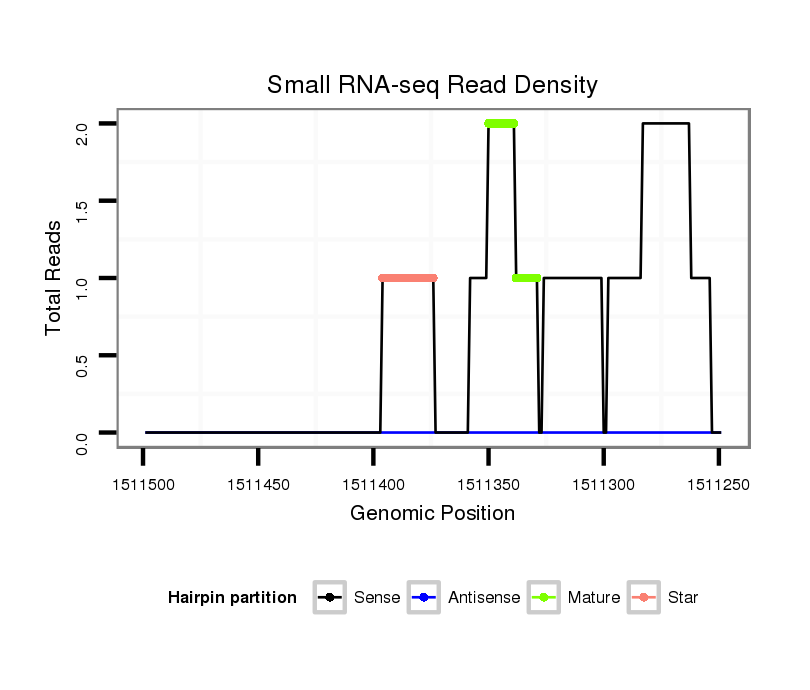
ID:dvi_14276 |
Coordinate:scaffold_13042:1511299-1511449 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
CDS [Dvir\GJ15725-cds]; exon [dvir_GLEANR_16162:1]; intron [Dvir\GJ15725-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CCGTGCAGATGCACGCCAGACGCAGCACGGCGGGCGTCAAACGGAAGCCGGTGAGGCGTTTGAGGAAGGATAATGTGATGCCAAAGCGAGGACGAAGGAAAACAAAAGCTGGCCGTAAAACCCCGACGACCCGGTGCAGCCAAACAGTCGAGGGTAACCAGGCAGGCAAAACTATGTTGTAGGAATAACTGGGCGAGGGGTAACAATGTGACAGGCGTGAGCGAAGGATTTAAATAGGAATTTACGTTTTG *******************************************************************************************************....(((.(((.....((((((((.....(((...))).....)))).)))).....))).)))....******************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060659 Argentina_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060683 160_testes_total |
M047 female body |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060658 140_ovaries_total |
M027 male body |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................GAGGGTAACCAGGCAGGCAAAA................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAACAGTCGAGGGTAACCAG.......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................ATGTTGTAGGAATAACTGGGCGAGGG.................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GTTTGAGGTAGGGTAATG................................................................................................................................................................................ | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTGAGCGAAGGATTTAAATAG.............. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GCGAAGGATTTAAATAGGAATTTACG..... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AAAAGCTGGCCGTAAAACCCCGA............................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACAATGTGACAGGCGTGA............................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CGAGGAAGAAGGAACACAAA................................................................................................................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................GACGCAGGACGGCGGGAG....................................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AATAGTCCAGGATAACCAGG......................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................GGTCAAACGGAAGCCTGTGC..................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................GACGCAGGACGGCGGGAGG...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................AACCGTAAGCCCGTGAGG................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................TAGGGATAACAGGGCGAA...................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GGCACGTCTACGTGCGGTCTGCGTCGTGCCGCCCGCAGTTTGCCTTCGGCCACTCCGCAAACTCCTTCCTATTACACTACGGTTTCGCTCCTGCTTCCTTTTGTTTTCGACCGGCATTTTGGGGCTGCTGGGCCACGTCGGTTTGTCAGCTCCCATTGGTCCGTCCGTTTTGATACAACATCCTTATTGACCCGCTCCCCATTGTTACACTGTCCGCACTCGCTTCCTAAATTTATCCTTAAATGCAAAAC
********************************************************************************....(((.(((.....((((((((.....(((...))).....)))).)))).....))).)))....******************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR1106720 embryo_8-10h |
V047 embryo |
SRR060682 9x140_0-2h_embryos_total |
SRR1106727 larvae |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................CTGCTGGGCCACGGCGGT............................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TGTTTTCGGCCAGCATTTTCGG................................................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TCAGCTCCGATTTGGCCGTC...................................................................................... | 20 | 3 | 7 | 0.43 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| ................................................................................................TCTGTTGTTTTCGACCGACATT..................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CCGACCGGCATTTTGGGC............................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CTACGGTTTGGCTCC................................................................................................................................................................ | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................CGCCCGCAGTTTCCCGTCCGC......................................................................................................................................................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................CAGCTCCGATTTGGCCGTC...................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:1511249-1511499 - | dvi_14276 | CCGTGCAGATGCACGCCAGACGCAGCACGGCGGGCGTCAAACGGAAGCCGGTGAGGCGTTTGAGGAAGGATAATGTGATGCCAAAGCGAGGACGAAGGAAAACAAAAGCTGGCCGTAAAACCCCGACGACCCGGTGCAGCCAAACAGTCGAGGGTAACCAGGCAGGCAAAACTATGTTGTAGGAATAACTGGGCGAGGGGTAACAATGTGACAGGCGTGAGCGAAGGATTTAAATAGGAATTTACGTTTTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/16/2015 at 11:06 PM