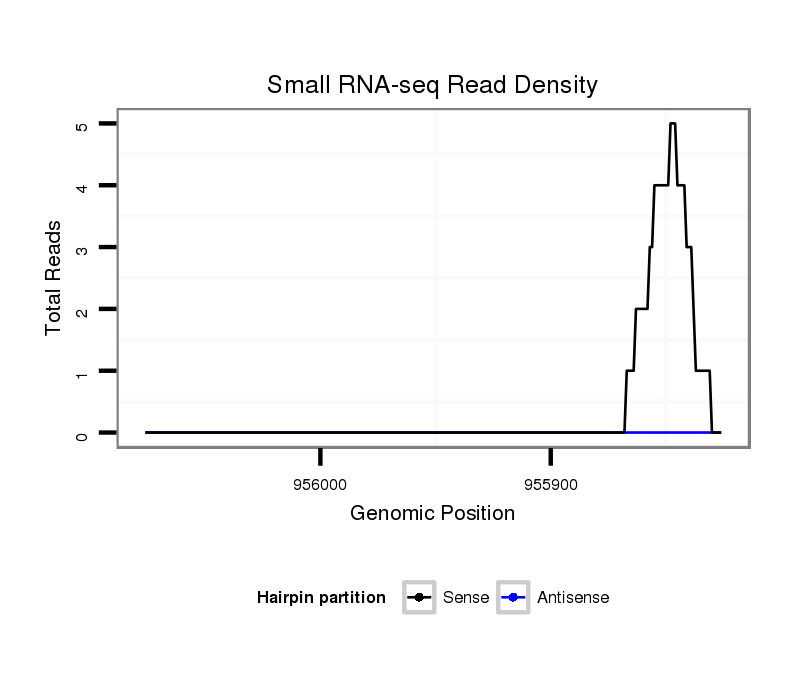
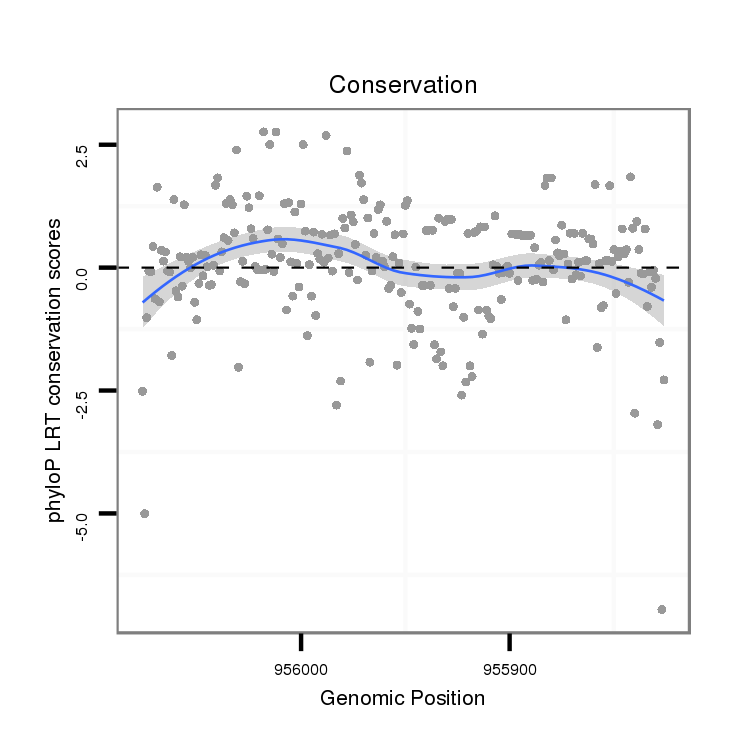
| droVir3 |
scaffold_13042:955826-956076 - |
dvi_14222 |
CGTG-TTTT--GTTTTTGTAG-----------TCGTTGTTTATA-TGT--------CACA-------CACA--CACAC--ACATA------CACGCACACTCACAC----GC--ACAC--------TCATACAA--ATAAGCACACGCAGAGAC---------------AGACAGAC------------ACA--G-----AGACA--AG---CGACAG--A----GACAAGCCAC------GAACCA---------------------C-GGACCAGACC--------------------------------------------------CAACGCC-------TCC-----------GATTGACACATTGGATTA---------------------------AATGCCCGAC--------GGCCC-A----TTACAGATCCAAAAGAATAGGCAAATGAT-GGAGCAGCTGGAGAA-GGATTG----------CAGCG------------A |
| droMoj3 |
scaffold_6496:21182139-21182342 - |
|
CA------------------------------------------------------CA---------CACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CAC--ACACACACACACACACAC---------------ACACACAC--------------ACAC-----AGGCAGCAG---CAGCAG--C----AGCA----------------------GCAGCTAAAGCCAATTGGTTGGGCAAAGTTGAGCGTCGCGGGTTAATTACCATATCAAATTCAAA-----------------------------------------------------------------------------------------------------------------------CAGAAACAAAGGCAACGG----------CAGCAGCTA--CAA-CAACAA----------CAACA------------A |
| droGri2 |
scaffold_15081:2081125-2081372 - |
|
TGTG-TTGTTTATT-TTTTAATTGGTAGT-AGATGTTGTCCGTAATCCCGCACCAAAACA-------TACA--CACAC--TCGCA------CACACACACACATAT------------G--CATACACACACAT--ATATACGCATAAAATATC---------------ACACACACACACACACTCACACA--CCAGCGAGACA--AT------------------------------------------------CG-ACTCACTG----------------------------------------------------------AC------------------C--------------------------------------------------------------------CGACTCGACCCAA----TTGCAGATACAGAAGAATAGGCAAACGAT-GGAACAGCTGGACAA-GGACTG----------CAGCGAGATTAATCCGGA |
| droWil2 |
scf2_1100000004590:1208038-1208197 - |
|
TTGG-TTAT--ATCATAC--------------------------------------CACACTACACACACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACG--CA--------CAC--ACACACACACACACGCAC---------------ACACACAC--------------ACA---------------CAC----ACACA------------------------------------------C---------------------------------------------------ACACACACACACACACACCCAC-------ACC-TA--------------CACATCAAATTAAGCCTTGTGGCT--------------------------------------------------------------------------------------------------------------C------------A |
| dp5 |
XL_group1a:2555546-2555692 + |
|
CG------------------------------------------------------CA---------TACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CGC--GCGCACGGACACAGAGAC---------------AGACACACACACAC--------ATAT----------------------------------------------------CCCATACCCCAT-ATCCAGTC----------------------------------------------------------------------------TC------------------------------------ATTTCTCAAGGACAAAACGGAAGTGCCCGG---CGATCCAG-------------------------------------------------------------------------TCC------------A |
| droPer2 |
scaffold_14:2039646-2039832 - |
|
CA------------------------------------------------------CA---------TTCACCA--ACACGC--A------CATAGATATACTCAC-CCGAT--ATACA--TA--------CGT--TTACAAG----T-----TAACCA-GACCCAACT--------------------------------G-----------------------TA--------------------CCTACACATTAG-ACCCAATG----------------------------------------------------------ACAATTTTCAATTGAATGCCC-C---------------------------------------------------------------CTGCGCCCTGACCC-A----TTGCAGATACAAAAGAATAGGCAGCATAT-GGAGCTGTTGGAGAA-GGAGTG----------CGACG------------C |
| droAna3 |
scaffold_13117:4185609-4185774 - |
|
CT------------------------------------------------------CA---------TACAAAC--ACACAC--A------TACAGAGAGACAAAG-AG-AG--AGAGA--GA--------CAA--AAATATATATATCAAGA-------------------------------------------------------------AAAG--A----TACAGTGCAAAAATCC------TAGATACATTAT-ATATAATG----------------------------------------------------------------------------CG------------------------------------TGTTATAGGTTA------------------AAAAAAATAAATAC-AAACAATTGTAATT-----------------------ACCAGTCAGAAAA-A----------------AATC------------G |
| droBip1 |
scf7180000394714:115457-115577 - |
|
TGTT-GTTT--TTTTTTGTG-------GCATGTTGTTGTTTGATATGTCAAGGTTAAGCA-------CACG--CACAC--CT--A------CACCTACACACACAC----AC--ACAC--------TCGTACAAAAATATACACACGCA--TAC---------------ACACT------------AATACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G |
| droKik1 |
scf7180000299277:117401-117525 + |
|
CA------------------------------------------------------CA---------CACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CAC--GCTGGCACACACAT-------GAC---------AGGCAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAGAAGGGTTTTCTAGAGAG-CTAGCTTCAAAAATCGTGGAAAACCCAGGCCCAA |
| droFic1 |
scf7180000454073:1697586-1697771 + |
|
AG------------------------------------------------------ATCA--------TTACGC--GCACTC--A------CTAACAAACAC--ACCAC-CCATCCAC----A--------CAC--ACAATCACACACTT-------CACGAGCCAATT--------------------------------G-----------------------TA------------------------CGACTTGG-ACCCACTG----------------------------------------------------------ACAACATTCAATTGAATGCCCCC---------------------------------------------------------------CTGCACCCTGACCC-A----TTTCAGATTCAGAAGAATAGGCAGATGAT-GGAGCAGCTGGACAA-GGAGTG----------CGATT------------C |
| droEle1 |
scf7180000490751:1079463-1079638 + |
|
AT------------------------------------------------------ATCA-------CCCAC-C--ACACGC--A------CACGCACACACTCAC-TC-AC--ACCCA--CA-------ACAC--ATA-------------------A-GACCCGATG--------------------------------G-----------------------TA------------------------CGAGCTGGAACCCACTG----------------------------------------------------------ACAACTTTCAATTGAATGCCCCC---------------------------------------------------------------CTGCACCCTGACCC-A----TTTCAGATACAGAAGAATAGGCAGATGAT-GGAGCAGCTGGACAA-GGAGTG----------CGACT------------C |
| droRho1 |
scf7180000779097:6242-6367 + |
|
TGCG-TCGTC----TGCATAA-----------TTGG--CTAATG-T--------GAAACA-------CACA--CACAC--AAACA------CACACACATACACAC----AC--AAAC--------ACACACAAACACACACACACACA--CAC---------------ACATG------------CACACA--GCATGTGAACA--GG------------------------------------------------CC-ATG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CA------------C |
| droBia1 |
scf7180000302193:855921-856040 + |
|
CA------------------------------------------------------CA---------CACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CAC--ACACACACACACACACAC---------------ACACACAC--------------ACA---------------CAC--ACAC--ACA----------------------------------------C---------------------------------------------------ACACACACACACACACAGAAAC-------ACA-C--------------------------------------------------------------------------------------------------------------------------------------A----------CACCG------------C |
| droTak1 |
scf7180000413601:850-973 - |
|
CA------------------------------------------------------CA---------CAGACAC--ACAGAC--A------CACAGACACACAGAC-AC-AC--AGACA--CA--------CAG--ACACACAGACACACAGAC---------------ACACAGAC--------------ACAC-----AGACA----CAC----AGACA------------------------------------------C---------------------------------------------------ACAGACACACAGACACACAGAC-------ACA-C---------------------------------------------------------------------------------------AGA---------------------------------------------------------CAC------------A |
| droEug1 |
scf7180000409109:28518-28705 - |
|
A-------------------------------------------------------CA-------CACAC----------TGATTCACAACCACACACATACATAC-AC-AC--AAAAA--CA--------CCA--ACACAAACACACAA-----TTCA-GATCCGATT--------------------------------G-----------------------TA------------------------CGACTTGG-ACCCACTG----------------------------------------------------------ACAATTTTCAATTGAATGCCCCC---------------------------------------------------------------CTGCACCCTGACAC-A----TTTCAGATACAGAAGAATAGGCAAATGAT-GGAGCAGCTGGAGAA-GGAATG----------CGAGG------------C |
| droSim2 |
3r:11874387-11874505 - |
|
GTGTTCTGTTAGC-TCTCTCG-----------TCGTCCTGCGTG-GGC--------CTCA-------CACA--GACAC--GAACACTCA--CTCACTCACACTCAC-AC-AC--ACA--------------CAC--AC--ACACACACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAGAGGGGGGCAGGCACAGG------------------------------------GGCG------------A |
| droSec2 |
scaffold_0:17596661-17596862 + |
|
CA------------------------------------------------------TA---------AACACAC--CCACAT--A------CACCCATACCCACACCAC-TC---AAGTGGTC--------CAC--TCAGACACAAATAT-------TAC---------A-------------------ATG---------G----------CAGCGA--G----TACAATACAA----------------------TAAATCAACGACTCGGGCAAACT--------------------------------------------------CA-ACAGCTCGACGGCCTG---------------------------------------------------------------CTACAAATAAATGC-ATAAACTTCGAGTGTCAAACAACAAGGAAAGTAT-GAA--AGCTGGGCGA-GAAGTG----------GGAAA------------G |
| droYak3 |
X:9447088-9447291 + |
|
CA------------------------------------------------------CA---------CACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CAC--ACACACACACACACACAC---------------ACACACAC--------------AAAG-----GG-------CAT--GCAA--A----AACA-----------------------------------AAAA----------------------------------------------------------GAAAAACCAAATCGAAGGACC-CAAAAATCCCAACTAAAGCGGGAGGG-----------------------------------------------------------------GAGAGAGAGACGGACAGAGAGA-GGGAGAGCAAGGGAGAGGGCGA----------TAGAGAGAGGGAGAGCAG |
| droEre2 |
scaffold_4845:19638700-19638837 + |
|
TC-------------------------------------------------------A-------CACACACAC--ACACAC--A------CACACACACACACAC-AC-AC--ACACA--CA--------CAC--ACACACACACACACACAC---------------ACACACAC--------------ACA---------------CAC----ACACACACACACAGGCGCC------TCACAA---------------------T-GGCC------------------------------------------------CACACATACAACGAC-------ACC-----------GAATGGGG------------------------------------------------------------------------------------------------------------------------------------C------------A |