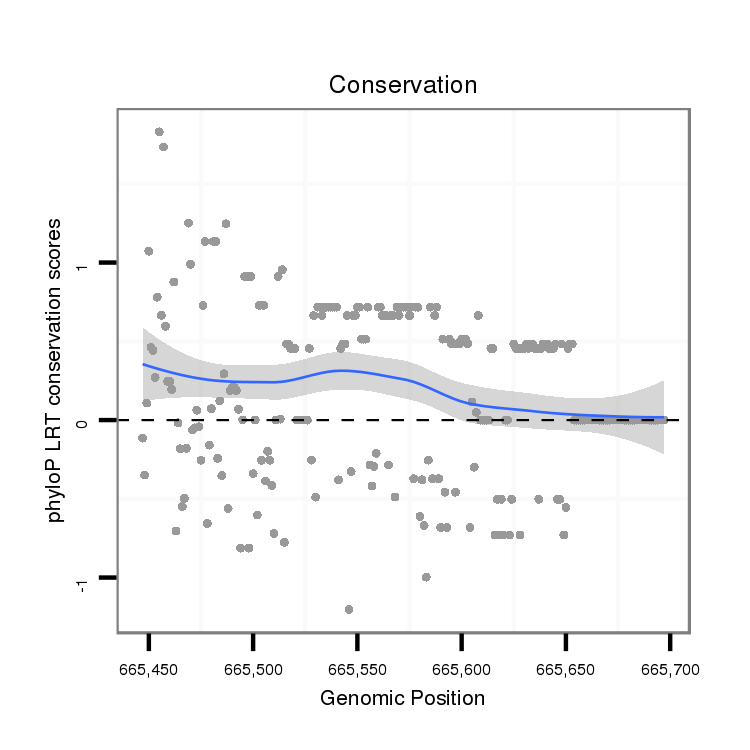
ID:dvi_14177 |
Coordinate:scaffold_13042:665497-665647 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_16256:1]; CDS [Dvir\GJ15818-cds]; intron [Dvir\GJ15818-in]
No Repeatable elements found
| --------##########################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACAAAACAATGTTGGGCGGAAGGGAGTGTGGTATGGGGAAGGAGGAGAGGGTGTGTGTGTGTGTGCAAGTAGGGCCAACCGAGGTTGATTAAAAAGAACGCCGAAAAAATTGAAACGCAGCTACAAAACAGAAGACGAAGCAGCAGCAGCGGCAAAGCATTAGACAAGCCGCGTAACATGCTCGACTAGCGGATACCCTATAAACAAAGCGATATTCGATTCGATATTTATCGATTTTCAATAATTCTATA **************************************************((......((((.(((.(((.((((....((.((((.............((((......(((....((.(((................))).))..))))))).............)))).))......)))).))).))).))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
V047 embryo |
SRR060666 160_males_carcasses_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|
| ................TGGAAGGGAGAGTGGTATGGG...................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................TGGAAGGGAGAGTGGTTTGGGG..................................................................................................................................................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .....................................GAAGGATGAGAGGAGGTGTGT................................................................................................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ...........................GTGGTATGGGGATCGAGTA............................................................................................................................................................................................................. | 19 | 3 | 14 | 0.14 | 2 | 0 | 2 | 0 | 0 |
| .........TTGTGGGCGGAAAGGAGTGT.............................................................................................................................................................................................................................. | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .........................................GTGGAGAGGGGGTGTGTGTGTC............................................................................................................................................................................................ | 22 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................GTAGAGCATGAGACAAGCC................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................AGTAGCAGCGGCAAAGCC............................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................CCGTGGTATAGGGAAGGA................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
TGTTTTGTTACAACCCGCCTTCCCTCACACCATACCCCTTCCTCCTCTCCCACACACACACACACGTTCATCCCGGTTGGCTCCAACTAATTTTTCTTGCGGCTTTTTTAACTTTGCGTCGATGTTTTGTCTTCTGCTTCGTCGTCGTCGCCGTTTCGTAATCTGTTCGGCGCATTGTACGAGCTGATCGCCTATGGGATATTTGTTTCGCTATAAGCTAAGCTATAAATAGCTAAAAGTTATTAAGATAT
**************************************************((......((((.(((.(((.((((....((.((((.............((((......(((....((.(((................))).))..))))))).............)))).))......)))).))).))).))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060681 Argx9_testes_total |
V053 head |
SRR060673 9_ovaries_total |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................CCCGGTTGGCTCCGACGAA................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TCGATCTTTTGTCTGCTGCTAC............................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................AAACTATAAATGGCTAAAAGT........... | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................CGATGTTTTGGCTTC..................................................................................................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:665447-665697 + | dvi_14177 | ACAAAACAATGTTGGGCGGAA-------GGGAGTGTGGT----ATGGGG-----AAGGAGGAGAGGGTGTGTGTGTGTGTGCAAGT----AGGGCCAACCGAGGTTGATTAAAAAGA-ACGCCGAAAAAA-------------------T--TGAAACGCAGCTACAAAACAGAAGACGAAGCAGCAGCAGCGGCAAAGCATTAGACAAGCCGCGTAACATGCTCGACTA-GCGGATACCCTATAAACAAAGCGATATTCGATTCGATATTTATCGATTTTCAATAATTCTATA |
| droMoj3 | scaffold_6308:477385-477576 - | GCAAAGCAATGTCGAGGGGGT-------GGGGGTG--GG----ATGGGTCAGCAAA--GGGGGCGGGGGCGG------AAGCAACTGCTAAGGG------GGGGTTGATTAAAAAGAAACAACGAA---AATAACAACGAAACGAGCAAC--AGAAACGCGGCGACAAAACAAAACAATGAACAA------------------------GCAAAAG--AGTGCGCGACTAGGCAGATACCCTGCACTCAA-------------------------------------------- | |
| droGri2 | scaffold_15203:5962920-5963040 + | TGGTTGG--------GAG-------------------GT----GTGGGG-----CAGC-------------------------------------------AGCTTGATTAAAAC----CCCCGAAAAAAAGAACAATGAAACGCGCAGCGAATGAACGCGGCGACAAAACAAAAGCAAAAACAAAAAAAGCAGCAAAGAAGTA------------------------------------------------------------------------------------------ | |
| droWil2 | scf2_1100000004909:9232600-9232606 - | ACGAAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | XL_group1e:7085275-7085293 + | ACAAAACAATGCTAGACA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_13:1468800-1468818 - | ACAAAACAATGCTAGACA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13417:1582157-1582227 + | ACAAAACAAAGCTAGACAATG-------AGGAAAGAGG------AGGAG-----GAGGAGGAGTAGGAGTAGGAGGAAAGACGAG-------------------------------------------------------------------------------------------------------------------TGGG------------------------------------------------------------------------------------------ | |
| droBip1 | scf7180000396621:315305-315357 - | AAA-------GCTAGACAATGAAGGAGGAGGAGAA--GAGAAAGTGGTG-----GAGGAGGAG-----------------------------------------------------------------------------------------------------------------------------------------AAGG------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000302495:262309-262334 + | ACAAGACAATGCTAGAGACAA-------G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGG------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000491272:1240614-1240614 - | A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000764255:78450-78471 + | GAAAAACAATGCTAGACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGG------------------------------------------------------------------------------------------ | |
| droTak1 | scf7180000415248:140116-140137 + | GAAAAACAATGCTAGACG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGG------------------------------------------------------------------------------------------ | |
| droEug1 | scf7180000409230:209494-209524 + | ACCAAAGG----CGAAGGATG-------TG-------------------------CGAAAAAC-----------------------------------------------------------------------------------------------------------------------------------------AATG------------------------------------------------------------------------------------------ | |
| droEre2 | scaffold_4690:10264124-10264145 - | AAAAAACAATGCTAGACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGG------------------------------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:17 PM