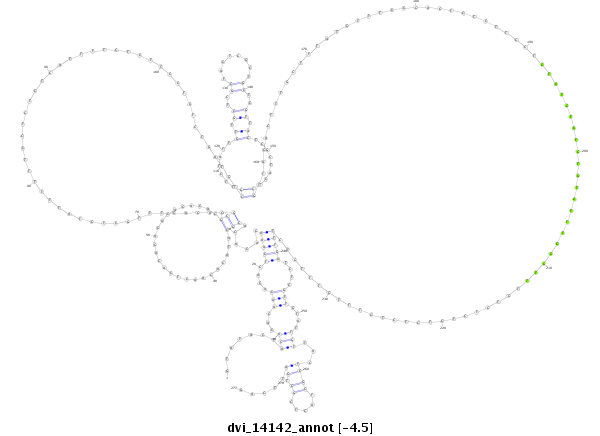
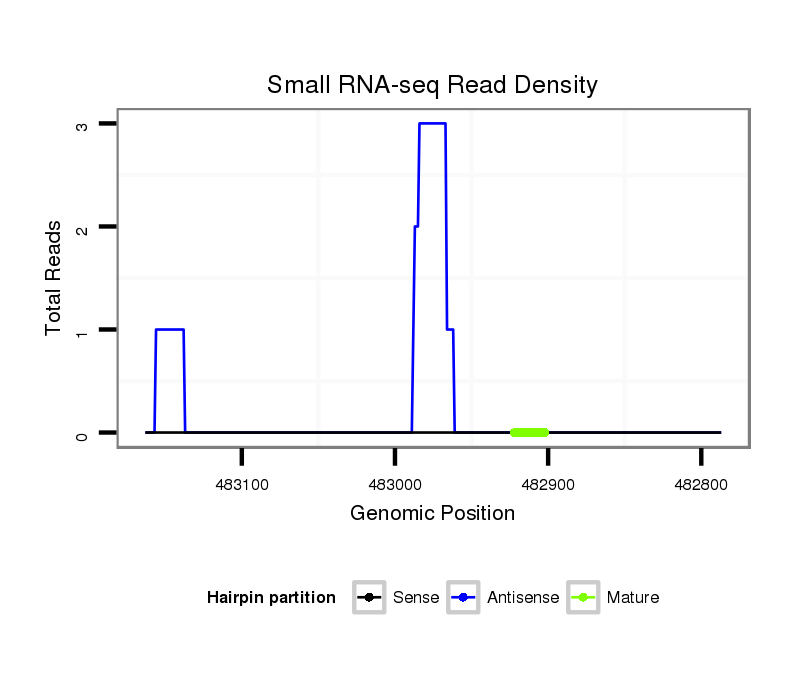
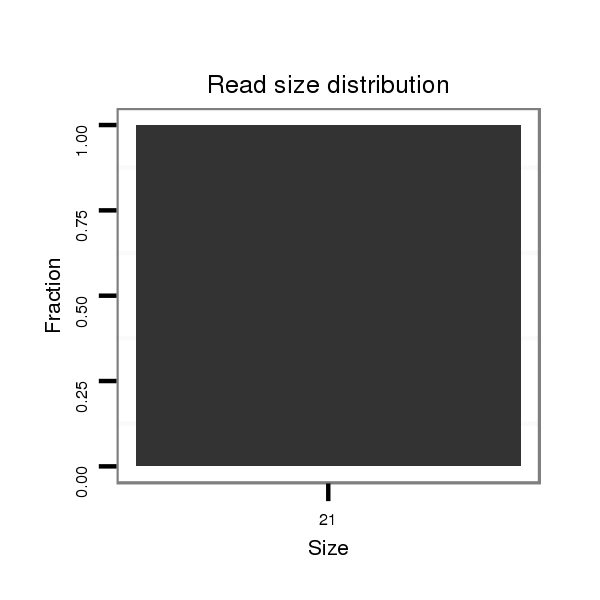
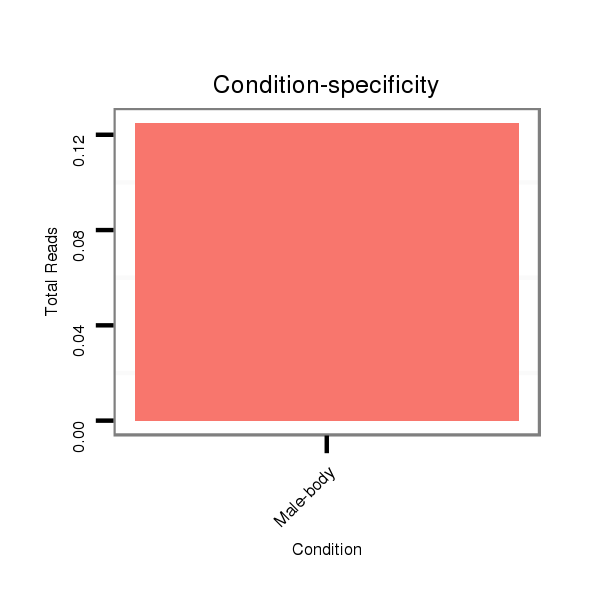
ID:dvi_14142 |
Coordinate:scaffold_13042:482837-483113 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -4.5 | -4.5 | -4.4 |
 |
 |
 |
exon [dvir_GLEANR_16221:5]; exon [dvir_GLEANR_16221:4]; CDS [Dvir\GJ15785-cds]; CDS [Dvir\GJ15785-cds]; intron [Dvir\GJ15785-in]
| Name | Class | Family | Strand |
| GA-rich | Low_complexity | Low_complexity | + |
| (TG)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACGAGACCGCCTCCCTCTCATCTGCCGACAATGATCCCGAGGATAGTCAGGTATACTACACACAACACACTCAAACACACACGCATACACAACTCACACACACACACACGCGCACACTTTGCTCTATTTTTTCCTCTCTCATTTTCTCTTCCTCTCTGATCTCGTTCTATTGTTGCTCGCTACTCGCTGCTCGTTACTCGCTACTCGTTACACTTTGCTTTATCTTTCCCACTCTCTTCCTCTCTCTCTCTCTCTTTCCCTTTCTCTCTCTCTCTCTTTCTTTCTCTTTGATCTCGTTCTATTGTTACTCGCTACTCGCTGATTCAGCACTCCTCTCTGCTCTCACTTACACAGGACTCGGTCTTCGAGGGCGTGCT **************************************************.......(((...(((....(((((......(((.........................)))..................................................((.......((.((..((........))..)).))........))...............................................................................)))))....)))....)))...((((.....)).)).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
M047 female body |
SRR060679 140x9_testes_total |
SRR1106729 mixed whole adult body |
SRR060664 9_males_carcasses_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................TTGCTCGCCACTCGCTGCTCGT....................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................AGTCAGCACTCCTCTCTGCTCTCACTT............................. | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TTTCTCTCCCGCGCTGATCTC...................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................CGAGGATAGTCAGGACT................................................................................................................................................................................................................................................................................................................................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................CTCTCTCTCTCTCTTTCCCTT................................................................................................................... | 21 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................CGAGGATAGTCAGGACTC.................................................................................................................................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................TTACATTTCTCTCTCTCTTTCTTT.................................................................................................. | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGCTCTGGCGGAGGGAGAGTAGACGGCTGTTACTAGGGCTCCTATCAGTCCATATGATGTGTGTTGTGTGAGTTTGTGTGTGCGTATGTGTTGAGTGTGTGTGTGTGTGCGCGTGTGAAACGAGATAAAAAAGGAGAGAGTAAAAGAGAAGGAGAGACTAGAGCAAGATAACAACGAGCGATGAGCGACGAGCAATGAGCGATGAGCAATGTGAAACGAAATAGAAAGGGTGAGAGAAGGAGAGAGAGAGAGAGAAAGGGAAAGAGAGAGAGAGAGAAAGAAAGAGAAACTAGAGCAAGATAACAATGAGCGATGAGCGACTAAGTCGTGAGGAGAGACGAGAGTGAATGTGTCCTGAGCCAGAAGCTCCCGCACGA
**************************************************.......(((...(((....(((((......(((.........................)))..................................................((.......((.((..((........))..)).))........))...............................................................................)))))....)))....)))...((((.....)).)).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
SRR060667 160_females_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060685 9xArg_0-2h_embryos_total |
V116 male body |
SRR060666 160_males_carcasses_total |
V053 head |
SRR060661 160x9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060673 9_ovaries_total |
SRR060658 140_ovaries_total |
M047 female body |
SRR060662 9x160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060664 9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................AGCGATGAGCGACGAGCAATG.................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GATGAGCGACGAGCAATG.................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GCGGAGGGAGAGTAGACGG............................................................................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AGAAGAAAAGGGGAGACTAG........................................................................................................................................................................................................................ | 20 | 3 | 11 | 1.00 | 11 | 0 | 1 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| ...............................................................................................................................................................................GAGCGATGAGCGACGAGCAATGAGCGA............................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAGAAAAGGGGAGACTAG........................................................................................................................................................................................................................ | 18 | 2 | 8 | 0.88 | 7 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAGAAAAGGGGAGACTAGTG...................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.36 | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................GTTGTGATAGTTTGTGTGTGC...................................................................................................................................................................................................................................................................................................... | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GAAGAAAAGGGGAGACTAG........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TGGAGAGACTAAAGCAAG.................................................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................GCGATGAGCAACGAGCTATGAGCGA............................................................................................................................................................................... | 25 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CTGTTACTAGTGACCCTATC........................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................AGAGAGAGAAAGAAAGAGAAA........................................................................................ | 21 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................AGCGATGAGCAACGAGCAATG.................................................................................................................................................................................... | 21 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................AGAGAGAGAGAGAGAAAG....................................................................................................................... | 18 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TGTGTTGAGTGTGTGTGT................................................................................................................................................................................................................................................................................. | 18 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ATATTGAAAGGCTGAGAGAA........................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................AGGAGAGGGGTGTTACTAG..................................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................AGAAGGAGAGAGAGAGAGAG........................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................................................................................AGAGAGAGAGAAAGAAAG............................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AAAGAGATAAGAAAGGAG................................................................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................AAAGAGAGAGAGAGAGAA................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GTGTTCGTATGTGTTGTGTG........................................................................................................................................................................................................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_13042:482787-483163 - | dvi_14142 | ACGAGACCGCCTCCCTCTCATCTGCCGACAATGATCCCGAGGATAGTCAGGT----ATACT-------ACACACAACACAC--TCAAAC--ACACACGCATACA---C-----------------A---ACTCACACACACACACACGCGCAC--ACTTTGCT---------------------------CTA----------------------------------------------------------------------------------------TTTTTTCCT-CTCTC-------------ATT---TTCT----------------------------------CTTCCTCTCTGATCTCGTTCTATTGTTGCTCGCTACTCGCTGCTCGTTACTCGCTACTCGTTACACTTTGCTTTATC------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCC--------C-AC----T--------------------------------CTCTTCCTCTCTC-----T--CTCTCTCTTT--C--------------CCTTTCTCT------------CT--------------------------------------------------------CTCTCTC-TTTCTTTCTCTTTG---ATCTCGTTCTATTGTTACTCGCTACTCGCTGATTCAGCACTCCTCTCTGCTCTCACTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACAGGACTCGGTCTTCGAGGG--CGTGCT----------------- |
| droMoj3 | scaffold_6540:12187132-12187389 - | TGCTTTAC---------------------------------------------------------------------------AC------AAGAAACTGC---GCAT-----TCC--CATACACACACACGTACGCACGCATGC---------------------------GTTTG--------------------------TCTTAATATCT--------GTCTGTCTCCCT-----GTCTA---------TACCACAGTCT----------GTCTCTTCGGTTTTCT-CTCTC------------------------------------------------------------------------------------------------------------------------------------TCTCTCTCTCC--------------------------CT-----------------------------------------------------------------------------CTCT-----------TTCTCTC------------------------------TCGCTCTCAC-------------TCCCTCTCT-CTGT-------------------------------C-----T--CTCTCTCTTT--C--------------TCTTTCTCT------------CTCTCTCTCGCTCTCACTCCTTCTCT------------------CT--GTC--TGTCTGTCTCTCT-CACT----------------------------------------------CTGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------G-CTT---------------C----TCT----------------CTCT--CTCTCT----------------- | |
| droGri2 | scaffold_15203:5811585-5811923 - | ATGAGACAGCTTCGCTTTCATCCGCCGATAATGATCCCGAGGATAGTCAGGT----ATGCTCCCATGCA---------CAC--A--CACACACACACACACACAG-ACATAC---ACACACACACACACACACACACACACAGAC---------------------------ATACA----------------------CACAC---------A--------CAC---------------------------------------------------------------ACA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACACATACACTTACACAAATACACTTACACAAATACACATACACCTTTCTTT-------------TTCGTTTCGTTTTTCGTTT-CGTTGCTTTCGTCT-------------CGTTTTCTCTATTCGAAACTG-------------------------------TTCT-TGTTCT----------CGC------------TACTCGCTACTCGCTCGTTCT------------G-----------------------------------------------------------------------------------------------------ATTTAGCACTCCTCTCTGCTCTCACTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACAGGACTCGGTCTTCGAAGG--CGTCCT----------------- | |
| droWil2 | scf2_1100000004511:10234427-10234677 - | TTCGTTTCATC-----------TGATGGCAACAAAG-----------CAGCTGAACAACCC-------TCACCCCACACAC--ACACACACTCACACACACACA------------------------CACTCACACACACACACAGGAACACCCACTC-----------------------GTTTAGCCTTGCTTG--------------CTG--------ATTAAGT----------------------------------------------------------------------A--ATCAAAG-----T------ATTTGG---------------------------------------------------------------------CAGCTAATTTATCGCCACTT---------------------------------------------------------------------------------------------------------------------------TACCCCTTTCTC-TCTCTCT----------------------------------------------------C-TC----T--------------------------------CTCTCT----CTCTCT-CT--CTCTCTCTCTCTCTCTCTCTGTATCTCT--------------------------------CTCTT----TC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---------------T----TTA----------------GACT--TTCTAT----------------- | |
| dp5 | 4_group5:1769491-1769721 - | ATACATCTCTC----------------------------------------------------------------------TC--------TCTCTCCCAC-------TCTT-------------------------------------------------C-GTCTC------------------------------------------------------------------TCCCTCTATCTC---------------TCT----------CTCTCTATCTATCTAT-CTCTCCCTCAC-------TCT---CTTTCTC-----TTT----CTCTCTCTCACTC-CTCTC-----------------------------------------------------------------TCTCTCTC--------------------------------ACTCTCTC-TCACTC------------------------------------------------------------------TCA---------------------------------------------------------------CTCTCT-CTCTCACTCTCTCT------------------------------CTCTCTCTC--T-CTCTTA----------C--------------TCTCTCTCT------------------------CTTAC----TCTCT--CTCTCACTCTTACTCTCTCTCT-----------------CTCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-TCT---------------C----TCT----------------CTCT--CTCACT----------------- | |
| droPer2 | scaffold_1:304220-304448 - | ACTCTCTCTCT-C--------------------------------------------------------------------AC--------TCTCTCTCAC-------TCTC-------------------------------------------------TC----------------------------------------------------------------------T-----CTCATTCTC---------------T----------CTCTCACT-------------CTCTCTC-------ACT---CTTTCTC-----TCTCTCTCTCTCTCTCACTCTTTCTC-----------------------------------------------------------------TCTCTC----------------------------------TCTCTCTC----------------------------------------------------------------------------------------------------------------------TCTCTCTCTCTCTCTCTCT-----------CT--CTCTCTCTCT-CTCT-------------------------------C-----T--CTCACTCTTT--C--------------TCTCTCTCT------------CTCTCTCTCTCTCTCAC----T--------------CTTTCTCTCT--CTC--ACTCTTTCTCTCT-CTCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-ACT---------------C----TCT----------------CACT--CTCACT----------------- | |
| droAna3 | scaffold_13417:6764833-6765052 - | CTCTGTCTCTC----------------------------------------------------------------------TG--------TCTCTCTGTC-------TCTC-------------------------------------------------T-----------------------------------------------------------------------------GTC--T---------------------------------------------------------------------------------------------------------------------CTCTGTCTCTCTGTC-----------------------------------------TCTCTGTCTCTCTGTCTCTCTGTCTCTCTGTCTCTCTGTCTCTCTGTC----------------------------------------------------------------------------------------------------------------------TCTCTGTCTCTCTGT-------------CTCTCTGTCTCTCTGT-CTCT-------------------------------CTGT-CTCTCT----------GTC------------TCTCTGTCTCTCTGTCTCTCT------------GTCTC----TCTGT--CTCTCT--------------GTC--TCTCTGTCTCTCT-GTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCT-CTT--------------------GCTC--------------AGTCT--CAGTCT----------------- | |
| droBip1 | scf7180000396735:1035592-1035890 - | CAAACACACTC----------------------------------------------------------------------AC--------ATACAGAGACACAGGAC-----------------------ACACACACACACACATTTATGAGGC-----T------GAGTGTGTGTGTTTGCGTATC--------------CTTGAGCGTCA--------GTTGGGCGCCGG-----GCATG---------TTGCATGAGTT----------GTCGTTGACGCGTTGA-----C-------------ATT---TTTC----------------------------------TTCCCTGCCTGCTGAAGTCCTGTT-----------------------------------------GCCCCTCAATACCCACACACT--------------------------C-------GCACATACTCCCCACTCT-CT------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTCTCTA-----T--ATATTTT-----T--------------TTACAGTTT------------TTTCGTGTTTGCCTGAC----T--------------TATGCAATCG--TTA--GCAATCCTTTTC-TTTCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-TCT---------------C----CTC----------------GACT--TCCTTC----------------- | |
| droKik1 | scf7180000302485:26550-26806 + | CTCTCTCTCTC----------------------------------------------------------------------TC--------TCTCTCTCTC-------TCTC-------------------------------------------------T-----------------------------------------------------------------------------CTC--T---------------------------------------------------------------------------------------------------------------------CTCTCTCTCTCTCTC-----------------------------------------TCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTC----------------------------------------------------------------------------------------------------------------------TCTCTCTCTCTCTCT-------------CTCTCTCTCTCTCTCT-CTCT-------------------------------CTCT-CTCTGT----------CTC------------TCTCTGTCTGTCTGTCTCTCT------------GTCTC----TCTCT--CTCTCT--------------CTG--TCCCTCACTCTCT-GTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCC-CTCTTTACCTCTCGCGCTCTCACGCT----------------CTCTCTCGCTCCGACAAGATCTGCGAGGG | |
| droFic1 | scf7180000452868:444-640 + | TTCTTTCTTTC----------------------------------------------------------------------TT--------TCCTTCTTTC-------CTTC-------------------------------------------------C-----------------------------------------------------------------------------TTC--C---------------------------------------------------------------------------------------------------------------------TTCTTTCTTTCTTTC-----------------------------------------TTTCTTTCTTTCTTTCTCTCT--------------------------CT-----------------------------------------------------------------------------CTCTTCCT-------T----CCTTGCTTTCTTTCTTTCTTTCTTTCTTTCTCTCT-------------CTCTCTCTTCCTTCCT-TTCT-------------------------------TTCT-TTCTT--------------------------CCTTCCTTT--------------------------------------------------------CT------------TTCCTTC-TTTCTCTCTGTCTGGCTGTCTAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTC----------------TTTT--CTTTCT----------------- | |
| droEle1 | scf7180000491008:2858874-2859121 - | ACACACAC---------------------------------------------------------------------------AC------ACACAATTAC-------ATCCAACC--GAAACACACACACACACTCACACAGAC---------------------------ATAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAGCTCAATTGAATATGAAAAATCCCTTCCGTTCTGTTGTCATATATGCTGTTTCCGGTTTCCGCTAACCGCAACTCAAACGTCGTTGTCGTTGTCGTTGTCGTTGTCGTTGTCGTCGTCGTCGTCATCACATTTTGGTTAGGATTTTTCTTTTCGTTTTCCCCCCTTT---------------T----CTT----------------TTTT--TTTCCT----------------- | |
| droRho1 | scf7180000779876:91697-91944 + | AACACACACTC----------------------------------------------------------------------AC--------ACACATGTAC-------ACGTAA-G--GATACTCT-----CCACACACTCACACAGTGGCAGCTAC-------------------ATCCTTACA-ATC--------------CGCTTGCGTCAAATATTTTGTTGGCCCCGG---------------GGCAAT---------TGTTGTTGCTCCTCGCTGACATGCGAATAATGCTCGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTGGGATTTTTGCTCCAGCCGCAAAAAGGACTTGCTTCCGCTCTCTC-TCTCTTTCTCTC-----------------------------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCTCCACTTTTCACTCTCTCT-------------------------------------------------------------------------------------------------------------------------------------------------------------T---------------C----CTT----------------CTCC--CTCTCT----------------- | |
| droBia1 | scf7180000301925:7474-7713 - | TTGATTCTACA----------------------------------------------------------------------AT--------TTTCTCGCCC-------TTTT--------------------------------------------------------------------------------------------------------------------------TACCATTATAT---TGG------------TCTCACTTTCCAATTTTTTTGTGCGCACATCTCACTTGC-------ACTGCCCTTTCTC-----------TCTCTCTCTCACTC-CTCTC-----------------------------------------------------------------TCTCTCTC--------------------------------TCTCTCTC-TCTCTC------------------------------------------------------------------TCT---------------------------------------------------------------CTCTCT-C---------------------------------------------------------T--CTCTCTCT----C--------------TCTCTCTCT------------CTCTCTCTCTCTCTCTCCTCTTCTCTCTCTC---CTCTTTCTCTCT--CTC---CTCTGTCTCTCT-GTCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-T----------------------CTC----------------CCTC--CTCTCT----------------- | |
| droTak1 | scf7180000415873:386256-386509 + | TTCCATTTTCC---------------ATTGATGGTTAAGCGAACGGTC------------------------------TAC--A--TATACATACATACATACG------------------------GACCAACGCACACATGCAGGCACAT-----------------------------GTTTAGTCGTGTTTACAT-------------A--------GCA---------------------------------------------------------------AAGCGCGCACGA--AACGGGG-----C------CTTTCG--------TAACCC---------------------TCGCTTCTCACGCTG-----------------------------------------TCTCCCTC-------------------------------------------------------------------------------------------------------------------------T-----------TTCTCCC------------------------------CCACCCCCAC-------------TTGCTAACT-CACTCTCTCTCTCTCTCTCTACTCTCTCTCT--CTCTCT-CT--CTCTCTCTCTCTCTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TC---------------T----CTG----------------TCTC--TCTCTC----------------- | |
| droEug1 | scf7180000409183:1026509-1026724 + | CATGTATTTGG-C---------------------------------------------------------ACTTAACACAC--TTCTAC--CCACTAACACACA---C-----------------T----ACCACACACAAAAATACAACCACCCACTCGGCT----------------------------------------------------------------------T-----TTGATTTCG---------------C----------CTCTTACT-------------TTGGCT---------------TTGTTC------------------------------------------------------------------------------------------------------------TCGCCC-------------------------------------------------------------------------------------------------------TCGCCCTCTCCT-TCCCTCT----------------------------------------------------C-TC----T------------------------------------CTCTCTCTCTCT-CT--CTCTCTCTCTCTCTCTCTCTATCTCTCT--------------------------------CTCTT----TCCGT------------------CTTTATAGCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-CCT---------------C----TCT----------------T-TC--TATTGT----------------- | |
| dm3 | chrUextra:28818368-28818603 + | CTCTTCTCTCT----------------------------------------------------------------------TC--------TCTCTTCTCT-------CTTC-------------------------------------------------T-CTCTT------------------------------------------------------------------CTCTCT-----------------------T--------------------------------------------------------------------------------------------------CTCTCTTCTCTCTTCTCTCTTCGCTCTTCGCTCTTCGCTCTTCGCTC-----------------------------------------------------------------------------------------------------------------------------------------TTCTCT-------------CTTCTCTCTTCTCTCTTCT-CTCTTCTCTCTTCT-------------CTCT-TCTCTCTTCTC---TC-----------------------TT------C--TCTCTTCT----------CTC------------TTCTCTCTTCTGTCT------T--CTCTCTTC-TCTCT----TCTCT--CTT-----------CTCT--CTT--CTCTCTTCTCTC-----------------------------------------------------TTCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-CTT---------------C----TCT----------------CTTC--TCTCTT----------------- | |
| droSim2 | x:15621964-15622140 - | dsi_18338 | ACGAGACGGCGTCGCTCTCCTCGGCGGACAACGATCCCGAGGATAGCCAGGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAG------------CATTATTGCCAGCGTCCAGCAACCACATAGAACACTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACACAAAACCCACACCAC-ACCACACCGCACACATGCTTCCCCCAATCCTCCCGGACCTCTG-----------------------------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCACTCCTCTCTGCTCTCCCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 | scaffold_17:344337-344513 - | ACCAGACGGCGTCGCTCTCCTCGGCGGACAACGATCCCGAGGATAGCCAGGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAG------------CATTATTGCCAGCGTCCAGCAACCACATAGAACACTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACACAAAACCCACACCAC-ACCACACCGCACACATGCTTCCCCCAATCCTCCCGGACCTCTG-----------------------------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCACTCCTCTCTGCTCTCCCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | X:5647245-5647428 + | TTTCTTGTTAA----------------------------------------------------------------------TT--------TTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC--------------------------------------------------------------------------------------------------------------TATGCGGTCTTTACCTTCTACCTGTTCTCTTTCTTTTTCTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCT--------C-AT----T--------------------------------CTCTTTCACTTTC-----T--CTTTTTCTTTCTCTCTCTCTCTCTCTCTCTCTCTCA------------CTCACACTCACACTCACTTTTTAACG------------------T---------------------------------------------------------------------ACCTAGTACACCTCTTTACTCTTTTTTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T----------------ATGG--GAACCG----------------- | |
| droEre2 | scaffold_4784:18912350-18912573 - | TCGTTCTATTT-C--------------------------------------------------------------------TC--------TCTCTCTTTC-------TCTC-------------------------------------------------CC----------------------------------------------------------------------T-----CTCTTTC---------------TCT----------TTCTCTTTCTCTTTCT-CTTTC------------------------------------------------------------------------------------------------------------------------------------TCTCTCTCTCTCTCTCT------------------------------------------------------------------------------------------------------------------T----TCTCACTCTCTCTCTCGCTCTCTCTCTCTCTCTCT-------------CTCTCTCTCTCTCTCT-CTCT-------------------------------CTCT-CTCTCT----------CTC------------TCTCTCTCTCTCTCTCTCTCT------------CTGTC----TCCCT--CTCTCT--------------GTC--TCCCTCTCTCTTT-GTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCC-CTT---------------C----TCT----------------CTGT--CTCCCT----------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 08:54 PM