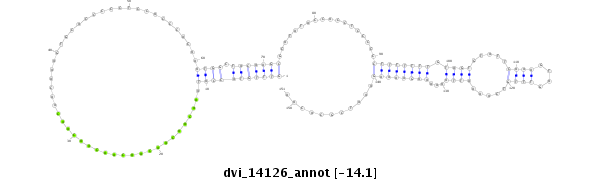
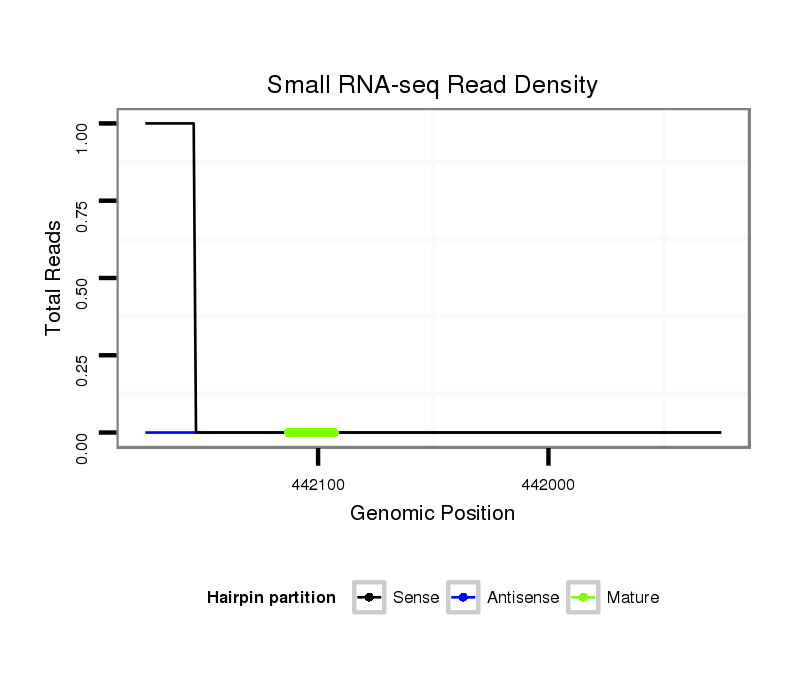
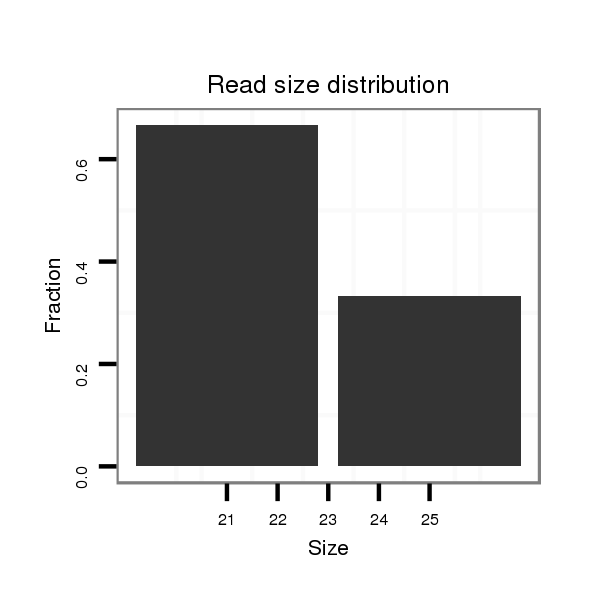
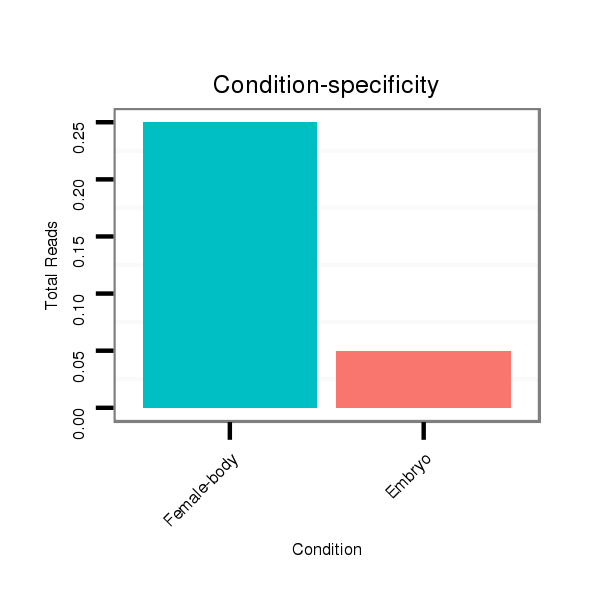
| droVir3 |
scaffold_13042:441925-442175 - |
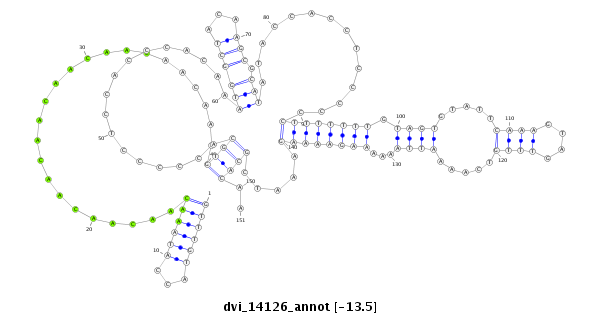
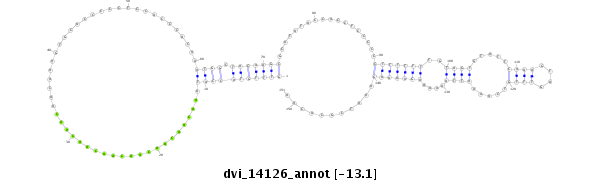
dvi_14126 |
AGACACTG------AGGATGA-GGATGG-TGATGGTATTGATAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGTCCGCTT-T-----TAGGTTTGTACCA-TA-----------------------------------------------------AACAACAACAACAACAACAACAACAAAGTGCACCCCCTCCACC---------------------------------------CA-CAAATGGCTACAAGCGC-----ATATACCACC---------TCCCCCCTTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGTAGTGTATTCAAAGTAG-----------------------------------------------------------------------------TTTGTCAAAAT-------------TAAAAAAGAAAAGAAAT---------------------CGCACAATATCTAACACTTGTTGTATATAAACACACACACT---TACACTTGCTAACTTG |
| droMoj3 |
scaffold_6359:1013323-1013532 + |
|
AGACACTG------ATGATGA-GGATGG-TGATGGTATTGATAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGTCCGCTT-T-----TAGGTTTGTACCA-TAAACAAC--------------------------------------------AAAAACAACAACAACAACAACAAC-ACAAACTGCACCC----ACCCAAAAACAATAAT---------ATACAAGAAC---------------------------------AACA---AC--------------AACTGTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGTAGTCT--------------------------------AACAAATGCACAAAACACTAAC---------------------------------------------------------------------------------------------------------------AA---ATGTATATAAAAA-----GCG---CACACTGTCTAACTTG |
| droGri2 |
scaffold_14853:4041376-4041645 + |
|
AGACACTG------AGGATGA-GGATGG-TGATGGTATTGATAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGTCCGCTT-T-----TAGGTTTGTACCA-TAAACAACAACAACAACAAA------------------------C----A---------------------------ACAAAGTGCACCCCC-----------------AAGCAACAGCA-AC----------------AATGGCAACAACTACAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCATTTATTTGTAGTGTATTCTACGTAGTT---------------------G----------TCCATTTTTTTTGCAAACATTTGC--TAACAAACCAAAAGAAAAACAAAAAAAAAAGAA-------------------------------GTTAAACAACAAAAACGC-CAAAAGTTAACA-TTATTGTATATAAACAC--------------------AACTTG |
| droWil2 |
scf2_1100000004909:11734564-11734801 + |
|
CATTGCAA------CAACA-A-CATCAA-CAACAATATCAACTG---CAA----TATAATAACAACA-----ACAACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AACAA------TAG----------------------------------------------C---------A-GCAACAGCAA------CA-ACAACAACAACAATCTC-----GA--------TGATGATATTTATTATTTATTTTATAAAGATTTTTATGTAAGTTTTCATTTAT----------------------------TTA----------------------------------AAATACAACAAAATAAAAATAAAAAAACATTTCTACTTCTTGCCCTCTCAATCTCTCACTCAAACATTT---------------------------------------------------------------------------------------ACATTTGC-------------------------------------------------------------------------------------------------------------------------T---GT------------CT |
| dp5 |
3:16501478-16501602 + |
|
AAAAAAAGTCAAAGCCAACAA-AAGAGGCCAGAG-------------C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-----TGG--------------------------------------------AGACCCAGCAAGACGC---A--TACAACAACAACAACAACAGCAACAAAAAGTACACCAACAACA---------------------------------------CG-AAAGTGGCGAAAGGTGC-----ACCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC------------CA |
| droPer2 |
scaffold_24:1415268-1415437 - |
|
TGAAACAA------CAACA-A-ATTCAA-TTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAAA-AG---------------ATAAAACA---------------GC----AACAGAAACAGCAACAGCAACAGCAACGACAAAACTC-----------------------------------------------AACTAA-AAAGTAACCAAAAAGTCA--GCAGAAAGAACC---------TCCTCCCCTCT----------------------------------------------ATCGAAACGGATAGAATA----------------------------------GAATATAAAAAAACAAAAACAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---AA------------GA |
| droAna3 |
scaffold_13417:199400-199569 + |
|
CGAGGAGG------AGCAGGA-GGATGGCTGGTGGTATTGATAG---AAA----AAAAAC-A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGCGCTTTC-C-----TAGGTTTGTACCA-CACACACA------------------C------ACACACACTCACACAC---ATACAGAACAACAACAACAACAACAT-AAACACTG----------------------------------------------CACTCA-AAAATATCCAAAAATTAA--AAAAAATATAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C---AC------------CT |
| droBip1 |
scf7180000395751:383182-383358 - |
|
GTAACAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAGCAACAGCAACAACAACCACAATCTGCAAAT----ACTG---------------------------------------CA-AAAGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTAAAGGTCGTTCGTTTCGAATTAATAGGATATTCATA-CAAATCATA--GAAAATCGCTTTAACTAAAC----------------AAT-------------TAAAAAACAAAACAAAAAATAAAGTT------------------------------------ATATAACTAAACAA----------GTGTCTTATTAA |
| droKik1 |
scf7180000302469:615452-615639 - |
|
ACACCAAC------AACA--T-CAACAT-CAACAACACCAACAC---CAA-----------------------CACTAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----AACAG------CAACACCAACAGC----------------------------------------------A-ACACCAACAACACCAGCA-ACACCAACACCAACACCACACCAACA---AC--------------------------------------------------------------------------------------------------AC----------------------------------------------------------------------------------CAACAAACACCAACAACACCAGTTACCAGTGCGTGGAATGTGTTG---------------------A----------AAAATT-----------------------------------------TGATTCAAAGGAACTTTTCG---------------------------------------------------------------------------------------------ACATT |
| droFic1 |
scf7180000454077:1466279-1466390 - |
|
AAGCACAA------CAGAT-A-TTGCAA-GT--------------------------------------AGCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCACA---------------------------------------------------------ACCACCACCACCGGCAACAGCAACAACAATTGC-------------------------------A-CCATCAGCAA------CA-GCAACAACAATTAGCAC-----CACCACCACC---------T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C---CT------------CC |
| droEle1 |
scf7180000491249:2550226-2550439 + |
|
AAATCCAA------ACCATGAAGGATCAACCGCAGCAACCCATG---GTG--AG-------------TC----GTCAATCGGTTTCGATTTTTCCCCTAAATGCCATTGACCTTTTCAG-------------------------------TTCTGCCC---CT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTACTCAATTAGTTTTGATTTTCATTTCATTTTCCGTTTCAAGGCAGCCAAATG--CAAAA--CACAAAATACAAAATACAAAACACAGAGCAAACCCATAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTCGCTTGGC---CA------------TA |
| droRho1 |
scf7180000779900:520116-520257 + |
|
AGCAGCAA------CAACA-A-CAACAA-CAACAACATCAAGTG---CAG------------CAACA-----ACAGCAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----ATCAG------CAT----------------------------------------------CAGCAACAGCA-AC----------------ATCAGCAACAACAACATC-----AG--------CAACAACA-------------------------------------------------------------------------------------------------------------------------------------------------------TCAACCTCAACCACCAACTCAACAACAGAAACCGTCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C---CT------------CC |
| droBia1 |
scf7180000301760:177345-177461 + |
|
GGCCGTCG------CCGATGA-GGATGGCTGGTGGTAGTGATAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAGTCGGCTTTTTGTTCTAGGTTTGTACCCACA-AA---------------ACA---CAGGCACAC------CCAC----A---------------------------AC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAGAC-----CC-----------------ACATCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAA------------------------------------------------------------------------------C---GA------------CT |
| droTak1 |
scf7180000415262:9017-9173 - |
|
AGACACAC------CTAAG-A-CATAAGCCGCTATCATTCACCC----------------------------------------TTAATCCC---TTTAAGGACACTTGACGTGTGCACGACCAACAACAACAACAGCAACAGCAGCAGGATCTGGCACACCTCCGGCCAAGTCGAAATGGCCATGTCCTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCC---------TGGCCCCTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTT |
| droEug1 |
scf7180000409061:268360-268563 - |
|
CGACAACA------ACAGCAA-CAACAA-CAGCAACAACAACAA---C-------------------TA----GACGAACAACTACAACACTTACAACA----ACAA-----------------------------------------GGACCTGGGAA---------------AACAGGAGCATAGCTTAGCTTTGGGCTGCTGTTGGCATACTTAAAGTTCATAAAAAAGTCAAATTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTCAAATTGCAATGCACACACACACAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACATGAACAACATACACACACACACA---CA------------CA |
| dm3 |
chrX:9080130-9080226 + |
|
AAGCAACA------ACAACAA-CAACAA-CAACAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACAAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTCTGTATTTTTTTT---------------------AAGCAAA------------------------------------ACCAAAAACAAAATATAAAAAAAAAAAACGAAAAA------------------------------------------------------------------------------A---AA------------CC |
| droSim2 |
x:4514067-4514158 + |
|
GGCTGTCG------CCGATGA-GGATGGCTGGTGGTATTGATAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAGTCGACTTTT-----TAGGTTTGTACCCACA-AAAACAACAACACAAAA------------------------C----A---------------------------ACACAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_0:3734781-3734924 - |
|
ATGACA--------CAGAATA-TTACTTCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAGCAGCAGCAGCAGCAGCAGCAAACTTCTTCC----TATC----------------------------AGCA--------------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCGACAATTGTTTGTTGTATCTTGCCACAGCGTTGAACTCAAACTCTG--AGCAGTTGC--TGA-------------------------AGAAACCGAAGAA------------------------------------------------------------------------------G---AA------------AA |
| droYak3 |
3L:11569271-11569417 - |
|
AGCCAGTG------CAGCA---CAACAACTGCTCAT--TAGTGATCGGAATT-----------------ATCCCAGCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATCAACA-------CCAGC-------------AACAACA---------------CC----AACAGCAGCAACAACACCAACAGCA-----------------------------------------------GCAGCAACAACAACCACA-GCGGTGGCCACAACATCATGTCACCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---CC------------TC |
| droEre2 |
scaffold_4690:2211320-2211444 + |
|
GGCTGTCG------CCGATGA-GGATGGCTGGTGGTATTGATAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAGTCGACTTTT-----TAGGTTTGTACCCACA-AA---------------ACAACACA---------------AC----A---------------------------AC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAGAC-----CCAACAAATTGTATGTTCCATATCCAAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGACTG------------CA |