

ID:dvi_13988 |
Coordinate:scaffold_12970:11097333-11097483 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
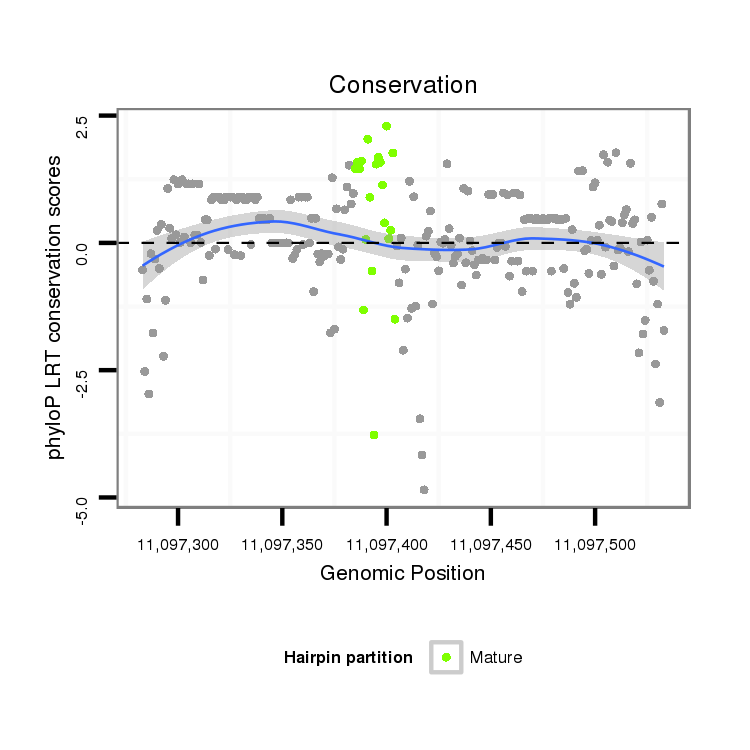
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.6 | -34.6 | -34.6 |
 |
 |
 |
exon [dvir_GLEANR_4414:1]; CDS [Dvir\GJ19525-cds]; intron [Dvir\GJ19525-in]
| Name | Class | Family | Strand |
| (TG)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTTACGACGAAAAAGAAGGATGCCGCCGCAGCCGACAAGGAGAATCTCTGGTGAGTTGAGCTTTAGCCTAGGCACAAGAAATGTGCATGCCTGTGTGTGTGTGTGTGTGTCTGTGTGTGTGCACTCAGCGTTTAATCATATGGCGCGCCTATTTTTCTTCTTTGTTTTTTTCTTGCTTTTAGGCAAATCAGCTGCTTTTGGCCAACTGCTTTTTGACTTTCTTTAGCTGACGTTTGCAATAATTTTCGGTT **************************************************...((((((..(((.(((((((..(((((((...(((.....((((((.((((..((((.(((.(((....))).)))))..))..))))..))))))..............)))...)))))))..))))))))))))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060663 160_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060681 Argx9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060654 160x9_ovaries_total |
M027 male body |
SRR060656 9x160_ovaries_total |
SRR060678 9x140_testes_total |
V116 male body |
M061 embryo |
SRR060673 9_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TTACGACGAAAAAGAAGGATGC.................................................................................................................................................................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AGCCGACAAGGAGAATCTCTGGTCTGC................................................................................................................................................................................................... | 27 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........AAAGAAGGATGCCGCCGCAGC........................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TTTACGACGAAAAAGAAGGATGC.................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GAAAAAGAAGGATGCCGCCGCA............................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CGACGAAAAAGAAGGATGC.................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................CAAGGAGAATCTCTGGTCTGC................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TACGACGAAAAAGAAGGATGC.................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGGATGCCGCCGCAGCCGACAAGG................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ACGACGAAAAAGAAGGATGC.................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TCTCTGGTGATTTGAGCTGTA.......................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TCTTTGGTGATTTGAGCTTT........................................................................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CGGACAAGGGGAATCACTGG........................................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TTAGTCTAGGCAAAAGAAA.......................................................................................................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TTTCTGTGTGTGCGCACTCTG........................................................................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......CGAGAAAGAAGGATGCTGCT................................................................................................................................................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................GTGTGTGTCTGTGTGTGTGC................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................ATGCCTGTGGGTGTGTGTGT................................................................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................AACGGAATCTCTGGTGAG.................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................TCTTATGAGTTGAGCTTT........................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
AAATGCTGCTTTTTCTTCCTACGGCGGCGTCGGCTGTTCCTCTTAGAGACCACTCAACTCGAAATCGGATCCGTGTTCTTTACACGTACGGACACACACACACACACACAGACACACACACGTGAGTCGCAAATTAGTATACCGCGCGGATAAAAAGAAGAAACAAAAAAAGAACGAAAATCCGTTTAGTCGACGAAAACCGGTTGACGAAAAACTGAAAGAAATCGACTGCAAACGTTATTAAAAGCCAA
**************************************************...((((((..(((.(((((((..(((((((...(((.....((((((.((((..((((.(((.(((....))).)))))..))..))))..))))))..............)))...)))))))..))))))))))))))))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060689 160x9_testes_total |
M047 female body |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060678 9x140_testes_total |
SRR060675 140x9_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................TGATCCTCTTAGAGACAAC...................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................CGGCGTCGGCTGTCCTTCTA............................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................AAAAGAAGAAACAAAAAA................................................................................. | 18 | 0 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ACGACAAACTGAGAGAAA........................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| .................TCTACGGCGGGGTCGGCT........................................................................................................................................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCGACTGCAAATGTGATTAA....... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................ACGTACGGACACACACACAC.................................................................................................................................................... | 20 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................GAAACCGGTTGAAGAAAA...................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:11097283-11097533 + | dvi_13988 | TTTACGACGA----AAAAGAAGGATGCCGCCGCAGCCGACAAGGAGAATCTCTGGTGAGTTGAGCTTTAGCCTAGGCACAAGAAATGTGCATGC--CT--GTGTG--TG------TGTGTG--TGTG---------------TC--T---GTGTG--------------------------------------------------------------------------------T----GTGCACTCAGCG---------------------------------------------------------TTTAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-------------------------ATATGGCGCG-------------------------CCTATTTTTCTTCTTTGTTTTTTTCTTGCTTTTAGGCAAATCAGCTGCTTTTGGCCA--------------------------------------------------------------------------------------------------------------------A------C-----------------------TGCTT-----------TTTG---------ACTT---------------TCTTTAGCTGACGTTTGCA-------------------------------------------------A--TA------A-TT--T--TCGG------T----------------T |
| droMoj3 | scaffold_6473:10264379-10264728 - | TTTACGACGA----AGAAGAAGGAAGCCGCCGCCGCCGACAAGGAAAACCTCTGGTGAGTTGAGCT---------GAGTAAGAA-TCTATAAATCTAT--GTGTATGTGTGTGTGT--GTGAGT-------------------------G---TG--------------------------------------------------------------------------------T----AGGGACTG----------------------------------------------------------------AGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-------------------------ATATGACGAG-------------------------CAAAT---------------------TGCGCTTGGCCTAAACAGCTGCTATTGGCTAAGAATTTTATTTATTTATTTTTTTTATATAATATATTTGTTATTTATTTTTGTTTTTAAATTTAATTTTTCTATTTATTTATTTTTA---TTTTTTTGTTAT------TTATTTTTATTTAATTTTGTTGCATTTATTTATTTATTA----------------TTTT---------ACTT---------------TCTTTAGCTGACGTTTGCA-------------------------------------------------A--TA------TATT--TTTT--T------T-----------GGC--A | |
| droGri2 | scaffold_15203:10352019-10352189 - | GCCC-AGCAA----ATAT----------------------------------------------------------------------GTGTGCCTGT--GTGTGTGTG----------------TGTG-------TGTGTGTG--T---GTGTG--------------------------------------------------------------------------------T------GTGTGT--------------------------------------GTGTATGTG----------TGTGTGTGTG-------------TGTGTGTT---T------------------------------------------------------------------------------------------GATTCTGTGTACGTCCCTTTTTTGGG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGTTTTGCGCCGGATAATAATAATCAGCGCGCTCCAATGCATACG---G--CT------AGTT--T--TTTG------T----------------T | |
| droWil2 | scf2_1100000004963:1126024-1126186 - | AAGG-------ATTGT--------------------------------------------------------------------------------GT--ATGAG--TG----------------TGTG-------TGTGTGTG--T---GTGTG--------------------------------------------------------------------------------TGTGTG------------------------------------------------------TGTGTGTGTGCTTGTTTCTGTGT----GTGTAT--------------------------------------------------------------------------------------------------------------------------------TTTTTCTTTTTTTT----------TTTTTGTTTGCTTCGGAACCAATGGCATTTTTCAAAAGTGACATT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGC--------------------------------------------GTG----CGCT---------------------------------------------T--TTGCTCT---TG--T--TTTGTTACAGT----------------C | |
| dp5 | XL_group1e:3378343-3378538 - | TTTACAACAA----AGAAGAAGGATGCCGCCGC---GGAAAAGGAGAATCTGTGGTAAGT---------------GCACAAAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTAAACTCTAATTGTTGGATA------------------------------------GTAGAT-------------TTT--TTCC-----------------------------------------------CTGTCCC----------TCTCTCTCGCGTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTCCTCTCTCGCCACTACTCTCTCCCCTGA------T-----------------------AAGTT-----------TTCA---------GCTT---------------TCTTTAGCTGACGTTTGCA-------------------------------------------------G--TA------GATT--TGTG--C------T----------------A | |
| droPer2 | scaffold_17:315597-315792 - | TTTACGACAA----AGAAGAAGGATGCCGCCGC---GGAAAAGGAGAATCTGTGGTAAGT---------------GCACAAAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTAAACTCTAATTGTTGGATA------------------------------------GTAGAT-------------TTT--TTCC-----------------------------------------------CTGTCCC----------TCTCTCTCGCGTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTCCTCTCTCGCCACTACTCTCTCCCCTGA------T-----------------------AAGTT-----------TTCA---------GCTT---------------TCTTTAGCTGACGTTTGCA-------------------------------------------------G--TA------GATT--TGTG--C------T----------------A | |
| droAna3 | scaffold_13047:104996-105145 + | TGTG-------TGTGT--------------------------------------------------------------------------------GT--GTGTG--TG----------------TGTA-------TCTG--TG--AGTG---TG--------------------------------------------------------------------------------T------GG-------------------------------------------------------------CTTATGTACTCTCATGG------TA----TT---ATT--TTTTCC-----------------------------------------------TTGCGCCCTATGAACAATTTTTGTTGCTTC-------------------------------------------------------------------------------------------------------------------TGGTTCGC----------TTTTAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCC------A-----------------------TTTTTGCGTACTCTCC---------------------------------TTCTGGTT---------------------------------------------------------------------------------TGTGTGTGT----------------G | |
| droBip1 | scf7180000396691:1703140-1703215 + | TACC-------TGTAT--------------------------------------------------------------------------------GT--GTATT--TGTGTGTGT--GTGTGT-------------------------G---TT------------------------------------------------------------------------------------------TTCAGCA---------------------------------------------------------TCGAAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-------------------------TTATGCTTTG-------------------------TTTACATTTCCCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTC------A----------------T | |
| droKik1 | scf7180000302592:520704-520908 + | TGCA-------GCCCT--------------------------------------------------------------------------------CT--GTGTG--TG----------------TGTC-------TGTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCT-------------CTC--TGCC-----------------------------------------------CTACCCC----------TCGCTCTCTCTCT--------------------------------------CTCTCTCTCTCTCTGTTTACATTGTTTACACGTATG------------------------CCACCGCTGCTGC-TGCTGGCCACCGTTCAACCGCTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGAATCCTTTTGCCTTCGTCCTTTTGGCG----------------TCTTTATTACAGCACTGCACTTAATAAACATTTGT---------------G-------------------------------------------------T--TA------A-A----TTT--C------T----------------G | |
| droFic1 | scf7180000454104:1167204-1167326 + | TGTG-------CATGT--------------------------------------------------------------------------------AT--TTACG--TG----------------TGTG-------TGTGTGTG--A---GTGTG--------------------------------------------------------------------------------TGTGTG------------------------------------------------------TCTATGCGAGAGTGTGCCACTTTGTGC------TG------------------------------------------------------------------------------------------------------------------------------CCTTCCTTTCTTTT----------ATTTTATTTTTTATAGAAAAA------------------------TCAATTTTG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C--T--CCTT------T----------------T | |
| droEle1 | scf7180000491272:342185-342302 - | TGTA-------TGTCT--------------------------------------------------------------------------------AT--GTGTGTTTGTGTGTGT--GTGTGT-------------------------G---TG--------------------------------------------------------------------------------T----TTGCAAGT--------------------------------------GTGCGTATGAGTG----------TGTGAG-------------TAAGCGTG---T------------------------------------------------------------------------------------------GTGTCTGTATTAGCCACTTTACT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTG------CTTATTGAACTGGGCAAA | |
| droRho1 | scf7180000779479:258832-258929 + | AACA-------GATAC--------------------------------------------------------------------------------GT--ATACC--TG----------------CG--ATGTGTATGTG--TG--T--G---TA--------------------------------------------------------------------------------T----TTGT------------------------------------TCGGTGG------------------AAAATGTCTTTTCGAAA------CG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCTTTTTG------------------------------------------TTTTATTTTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT--T--ACTT------T----------------T | |
| droBia1 | scf7180000301703:198096-198246 - | TGTG-------TGTGC--------------------------------------------------------------------------------GT--AGGGG--CG----------------CGTG-------TTTGTGTC--G---GTGTG--------------------------------------------------------------------------------T----GTGCACTG----------------------------------------GTGTGGG----------TGGCTGGGCG-------------AGTGTATT---GAA--CTCTTTCTGCATTTTTTGCACAATTTACCCGCTTTTGTTTTTTTTTCTTTTGCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCACT--GTA----------------------------------------------------------------------------------------TC-------------------------------------------------A--TT------G-TG--T--TCAT------T----------------T | |
| droTak1 | scf7180000415394:549805-549889 - | TTTT-------TTTGT--------------------------------------------------------------------------------TG--CTGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCTGCTACTTTTTTCGTGGC------A-----------------------TATTTTTTGACTTTGA---------------------------------CTTTAATT--CGTTTGCC-------------------------------------------------T--TTGCT---T-TT--T--TCCT------T----------------T | |
| droEug1 | scf7180000409006:261031-261189 + | GAAA-------TGTAT--------------------------------------------------------------------------------AT--ACATG--TGTGTG--TGTGTG--TTTG---------------TG--T---GTGTG--------------------------------------------------------------------------------T----GTGTACTGGTCG---------------------------T--------------------------------CATTCG----GTTCTT-----------C--TTCTTTTTGTTCATCTTCTTTGCAA--------------------------------------------------------------------------------------------------------------------------------------------------------------TGCATTTTGCTGCATTTTATTTATTGTTCCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGCTGGATGT------------------------------------------------------------------------------TGTTGCATGCA-------------------------------------------------T--AC------A-TT--T--TGCA------T----------------T | |
| dm3 | chr3R:9455149-9455308 + | GGCAAAAGAT----AA-ATATGCATGCGGTTGG----------------------------------------------------------------T--TT--ATGTGTGTGTGT--GTGTGT-------------------------G---TG------------------------------------------------------------------------------------------TTAGCCA---------------------------------------------------------TTTCAACAG----ACGCGC-----------C--TCCTTTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------------------------------------------CGCCGCCTTTCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCATTTGC------------------ATGCAATAAAGATGG------------------------------------------------CGGGGCGATAACTAGGTTGCCTGCCA--TTG-TTTGC--CTTT------T----------------C | |
| droSim2 | 3r:7038370-7038490 + | GC-G-----TGCGTG----------------------------------------------------------------------------------TGTCT--G--TG--------------TGTG---------TGAATGCG--AGTG---CGCTGGTGTTGAAAAA-----------------------------------------------------------------------------------------------ATC------------------------------------------TATCTCTTCGCTA------CGCATTTTCAC--------------------------------CCC----------------------------CC----------GTTTTTGCGTATTTGA--------------------------TCTCTCTTCCTCTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT--C--TCTA------T----------------C | |
| droSec2 | scaffold_21:353483-353650 - | CTTG-------TGTGT--------------------------------------------------------------------------------GT--GTGTG--CG----------------TGTG-------TGCGTGTTACCACC---CGAAAATGTTGCCCCAATCGACTTAATTGATTTGTGGGCATAACCAACACATATTTACACTCAGTTCGTATCTGCGTGTGTGT------GT------------------------------------GCG------------TTCT----------CATG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGATT-----------TACT----------------------------------GCTTACATATGCG-------------------------------------------------A--GT------G-TG--T--GCTT------T----------------T | |
| droYak3 | X:8048090-8048265 + | TGTATGTCGT----AT------------------------------------------------------------------------------------GTGTG--TA--------TGTG--TACG-----------TG--TA--C-----GTG--------------------------------------------------------------------------------T----GTGTCTTGAGCTTATACCGAAAAAAATTGTTTACAACTTTTGAAG------------------------------------------------------------------------------------------------------------------------------------------CT----------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTTTTAG---------------------------------------------------------------------------------------GTTTCCAATGTCCTGCTATTTGTTTACCTCATTACATTACTATATATAAATG------------------------------------------------------------------------------TGTGTAGATATCATATTCA-------------------------------------------------T--TA------A-CT--T--GACT------T----------------T | |
| droEre2 | scaffold_4690:1041725-1041885 + | TGTG-------CGTGC--------------------------------------------------------------------------------GT--GTGTG--TG----------------TGTGAGCTGTATCTG--TG--G---GTGTG--------------------------------------------------------------------------------T------GT------------------------------------GCACACAGATACAGG----------CTTATGTACT-------------CGTGTTTTTTC--------------------------------CCCACTTTTCTGTTTTTTTTTTTTGCTCGCGCCCTACGAAAAA---------------------------------------------------------------------------------------------------------------TTTGTTTTG-------------------------CTCTCAATGCCTT--------------------------------------------------------------------------------------------------------------------------------------------------------TTCTCCT-----------------------------------------------------------------------------------------------------------------------------------------------------------------CG--T--TTGG------T----------------T |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:15 AM