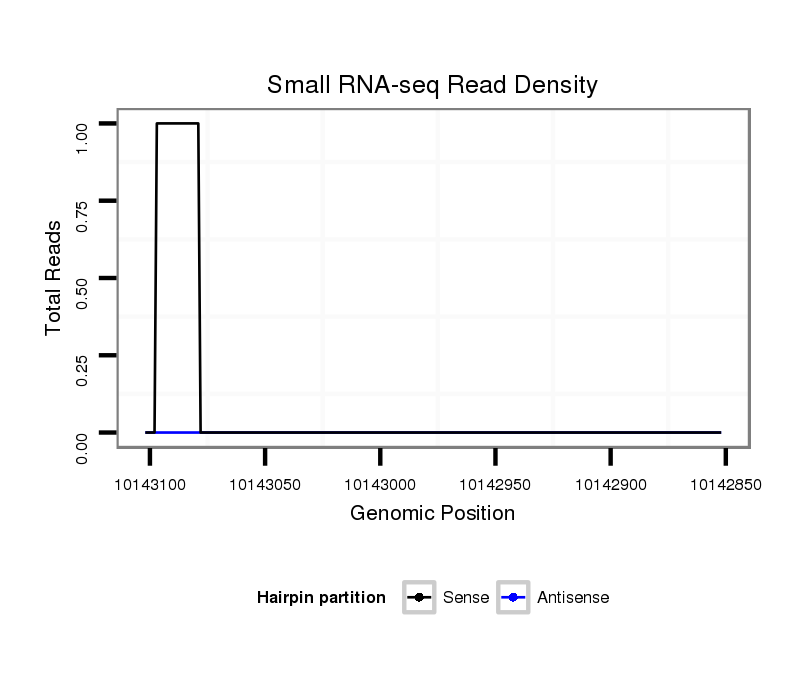
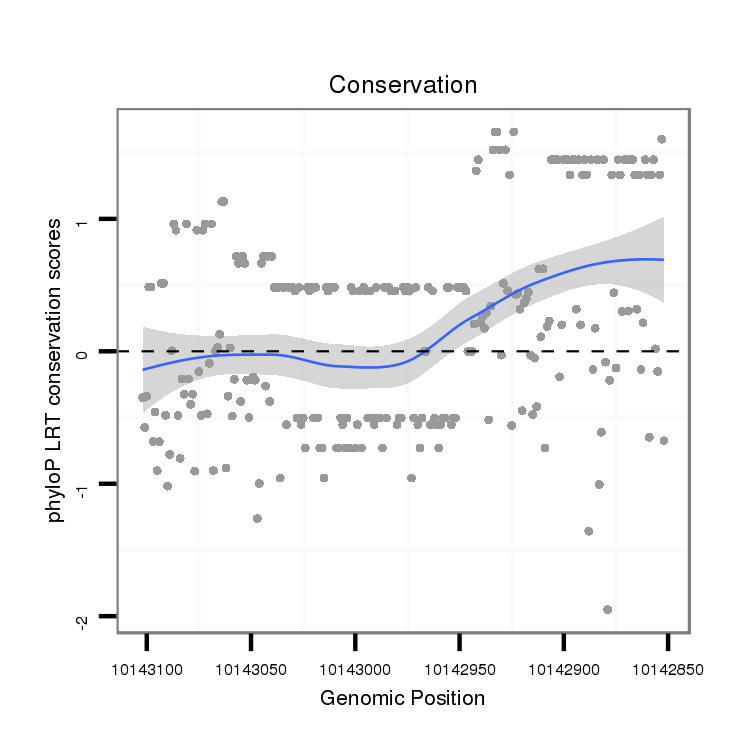
ID:dvi_13873 |
Coordinate:scaffold_12970:10142902-10143052 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18663-cds]; exon [dvir_GLEANR_3490:6]; intron [Dvir\GJ18663-in]
| Name | Class | Family | Strand |
| Helitron-1_DVir | RC | Helitron | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TACGCCGGATTGCGATGGTCTTCAAAGTGCTTAACTTTAAGCACAAGAAGGTAGGCCGATTATTTACTTATATGAATTGTCAATGTGCTCAATTGCAGGGACAAGCACTCGATAGCAGTGGGTGGTCAGCATTGGGCAAGAAGACATATGCATATGCAACAATGCATAGAAGAAGACTTTGTCAGCTTTGTAATTCATATTGATTCTTGAGCAGTATCAAGAAGCGCTTCGATTTAGCCCATCTGTTTGTG **************************************************...................((((((((((((....((((((((.((...(((...((((((.......))))))))).))))))))))....))))(((((((........)))))))((((..(((...)))..))))...)))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060679 140x9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
M061 embryo |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................AAGCACTCGATAGCTGTGGGT................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CGGATTGCGATGGTCTTCA................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGCAACAAAGCATTGAAGAAGACG......................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAAGACATTGTCAGGTTTGGA........................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAAGACATTGTCAGGTTGG............................................................. | 19 | 3 | 19 | 0.11 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...........................................................................................................................................................................................................................AGAAGCGCTTCGATGTCGGC............ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................AGTGGGATCAAGAAGCGCT....................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................TTAGATGAATTTTCAATGAGC................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ATGCGGCCTAACGCTACCAGAAGTTTCACGAATTGAAATTCGTGTTCTTCCATCCGGCTAATAAATGAATATACTTAACAGTTACACGAGTTAACGTCCCTGTTCGTGAGCTATCGTCACCCACCAGTCGTAACCCGTTCTTCTGTATACGTATACGTTGTTACGTATCTTCTTCTGAAACAGTCGAAACATTAAGTATAACTAAGAACTCGTCATAGTTCTTCGCGAAGCTAAATCGGGTAGACAAACAC
**************************************************...................((((((((((((....((((((((.((...(((...((((((.......))))))))).))))))))))....))))(((((((........)))))))((((..(((...)))..))))...)))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
SRR060686 Argx9_0-2h_embryos_total |
M061 embryo |
M027 male body |
M047 female body |
SRR060677 Argx9_ovaries_total |
V116 male body |
GSM1528803 follicle cells |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........ACGCTACCGGAAATGTCACGAA........................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................AATAGTTCTGCCCGAAGCTA.................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................CCAGAAGTTTCAAGAGTAGA....................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CTACCAGAAGGTTGAAGAA........................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GCGAAGCTAAATAGGGCAAA....... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................ACGAATTGAAATTCTTCCTC............................................................................................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......CTAACGCTGCCAGATGTTC................................................................................................................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................CTATCGTCTACCACTAGTC.......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................AATTCGTGTTCTGCC........................................................................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:10142852-10143102 - | dvi_13873 | TACGCCGGATTGCGATGGTCTTCAAAGTGCTTAACTTTAAGCACAAGAAGGTA---GGCCGATTATTTACTTATATGAATTGTCAATGTGCTCAATTGCAGGGAC-AAGCACTCGATAGCAGTGGGTGGTCAGCATTGGGCAAGAAGACATATGCATATGCAACAATGCATAGAAGAAGACTTTGTCAGCTTTGTAATTCATATTGATTCTTGAGCAGTATCAAGAAGCGCTTCGATTTAGCCC-A----TCTGTTTGTG-------------------- |
| droMoj3 | scaffold_6541:1057185-1057422 - | ACT---------------------------------TTAAATTCAAGAAGAGAAGAGAGAGAATATTTAGTTTTATGGAACTTCAGTAGTGTCAATGTTTTATACCCTGAACCCATTGAAAATGGGAAATCAGGGTATA--ATGGTTTTGTAAATATATG---TAACAGGCAGAAGGAGGCGTGGCAGACCCCATAAGGTATATATATTCTTGATCAGGACGACGAACTGAGTCGATTTAGCCCAG----TCCGTCTGTCCGTCCGTCTGTCTGTCCGTA | |
| droGri2 | scaffold_15252:8326047-8326119 + | TACGCAACCTTTGCATGTCCCTCTGCGCTCTGAAATATAATCTGGAGCAGGTG---AGATGATTCT-------------------------------------------------------------------------------------------------------------------------------CATTG--------------------------------AA-----------------------T--TG-------------------- | |
| droBip1 | scf7180000393226:11103-11162 + | ATT--------GTGGTGGGGTTGAAGGCGCTGCAGCTAAAGTT---------------------------------------------------------------------------------------------------------------------------------GAAGGTGA-GTTTCTGATCCTCTTAT---------------------------------------------------------------------------------- | |
| droKik1 | scf7180000301832:11458-11550 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACGCAATGAAGGAGACGTTTCCGACCTTATAAAGTATATATATTCTTGATCAGCATCAACAGCCGAGTCGATCTAGCCATG----TCCGTTTGTC-------------------- | |
| droEle1 | scf7180000490245:41597-41687 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACGCAGTGAAGGAGACGTTT-TGACCCTATAAAGTATATATATTCTTGATCAGCATCACTAGGAGAGTCGATCTAGTCC-G----TCTGTTTGTC-------------------- | |
| droRho1 | scf7180000778882:1440-1537 - | A------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACGCAGTGAAGGAGACATTTCCGACCCTATAAAGTATATATATTCTTAATCAGTATCACCAGCAGAGTCGATCTAGCCATGTCCGTCTGTTCGTT-------------------- | |
| droBia1 | scf7180000302060:21128-21219 - | A--------------------------------------------------------------------------------------------------------------------------------------------------------------------ACGCAGTGAAGAAGACGTTTCCGACCCCATAATGTATATTTATTCTTGATCAGCATCACTAAACGAGTCGATCTAGCCATG----TCTGTTCGTC-------------------- | |
| droTak1 | scf7180000414860:27595-27687 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACGCAGTGAAGGAGACTTTTCCGACTACATAAAGTATATATATTCTTGATCAGTACCAATAGCCGAGTCTATCTAGCCATG----TCCGTCTGTC-------------------- | |
| droEug1 | scf7180000409230:588595-588596 + | TA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/16/2015 at 10:57 PM