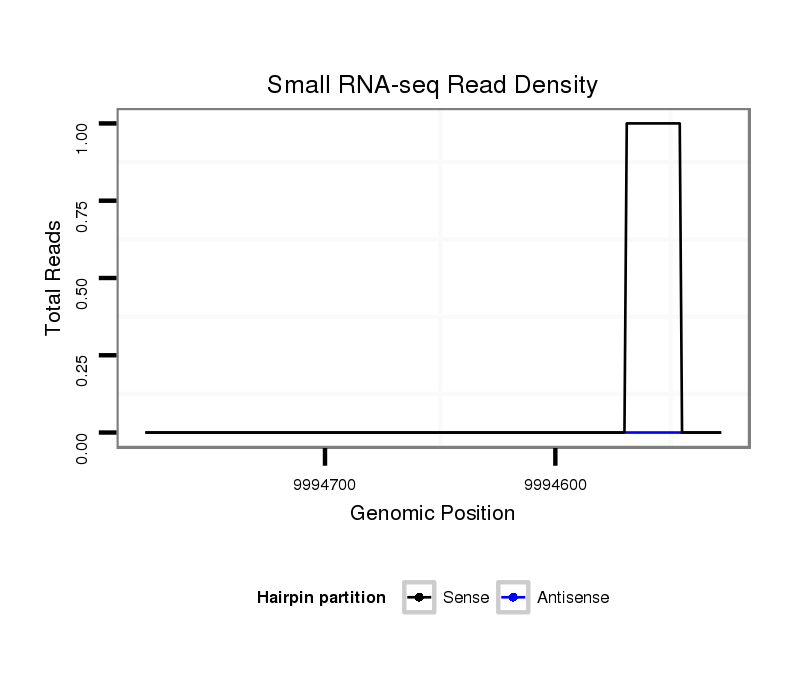
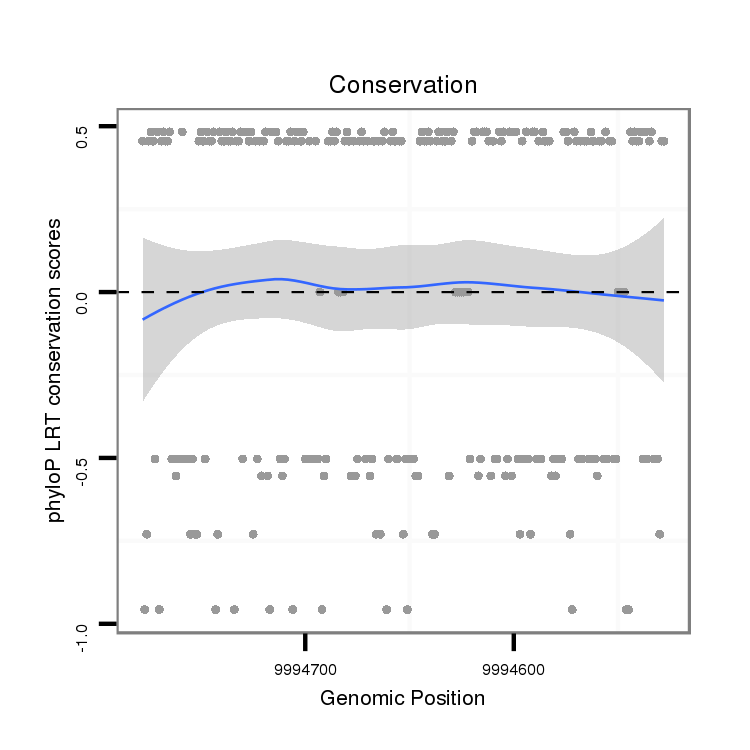
ID:dvi_13857 |
Coordinate:scaffold_12970:9994578-9994728 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18667-cds]; exon [dvir_GLEANR_3494:5]; intron [Dvir\GJ18667-in]; Antisense to intron [Dvir\GJ19479-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GGACTGTACCAGCAATATTAATGTTGAGAGTAGCTCAAGCAGCTCCCAAATGAACTCTGAAGTATCATCGCAGGATCTGCCAGGCTCTTGCTCAGCCGAGTCATCACGGTTCTCGCACTCAGCGCCAGTTGAAGAGGTGCTCATCGATCACTCCACTTCATTCATACACACAGTTAAATATAGTTTCATTCTTGCCTCTAGGATGACAGTCCCCAACCTGCTAAGGTACGCTCCTCAGGTAAACACTTGCG **************************************************((((...((((((......((((((.((....)).))))))..(((((........)))))..(((.....)))...(((((.(((...))).)))))......))))))))))................(((...((....))...))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060689 160x9_testes_total |
V047 embryo |
SRR060674 9x140_ovaries_total |
V053 head |
M061 embryo |
M028 head |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TCCCCAACCTGCTAAGGTACGCTC.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GATAACACTCCCCAACCTGCA............................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................AGCTCCGGTTGAAGAGGT................................................................................................................. | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TCGCAGGATCTGCCAGCCTCTTG................................................................................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TGCACCAGCAATATTAATGTTGAGAGTA........................................................................................................................................................................................................................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TTCATTCTTGACTCGAGGAAG.............................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................TCGGATGACAGTCGGCAACC................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................CGAGTCATCTTCGTTCTCGC....................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GGATGACAGTCGGCAACC................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................TAATACAGACAGTTAAATATG..................................................................... | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................TAATACACTCAGTTAAATAC...................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................CGAGTCATCGTGGTTGTCG........................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................GTTCTAGCATTCAGGGCC............................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CCTGACATGGTCGTTATAATTACAACTCTCATCGAGTTCGTCGAGGGTTTACTTGAGACTTCATAGTAGCGTCCTAGACGGTCCGAGAACGAGTCGGCTCAGTAGTGCCAAGAGCGTGAGTCGCGGTCAACTTCTCCACGAGTAGCTAGTGAGGTGAAGTAAGTATGTGTGTCAATTTATATCAAAGTAAGAACGGAGATCCTACTGTCAGGGGTTGGACGATTCCATGCGAGGAGTCCATTTGTGAACGC
**************************************************((((...((((((......((((((.((....)).))))))..(((((........)))))..(((.....)))...(((((.(((...))).)))))......))))))))))................(((...((....))...))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060655 9x160_testes_total |
V116 male body |
GSM1528803 follicle cells |
SRR060678 9x140_testes_total |
M047 female body |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................AGGTTGGACGACTCCATGC..................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................TCGAGTTCGTCGTGGGTTTA........................................................................................................................................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TCAATTTATATCAGAGTAAT............................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................TTACAACTCTTATCCAGTTCA................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................GTCGCGGTCAAGTGCGCCAC................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................TCAATTTAGTTAAAAGTAAGAA.......................................................... | 22 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ACCACGAGCGTGAGTCTCG.............................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| CCTGACTTGGACGTTAGAA........................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................GTCGTGGTCACATTCTCCA................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:9994528-9994778 - | dvi_13857 | GGACT----------GTACCAGCAATATTAATGTTGAGAGTAGCTCAAGCAGCTCCCAAATGAACTCT---GAAGTATCATCGCAGGATCTGCCAGGCTCTTGCT---CAGCCG-------AGTCATCACGGTTCT------CGCACTCAGCGCCAGTTGAAGAGGTGCTCATCGATCACTCCACTTCATTCATACACACAGTTAAATATA-----GT----TTCATTC----------TTGCCTCTAGGATGACAGTCC---CCAAC---CTGCTAAGGTACGCTCCTCAGGTAAACACTTGCG |
| droMoj3 | scaffold_6473:11527440-11527729 + | GCCCTGCCGATAGCAGCAGCAGCAGCTCCAGCAGCTCGAGCAGCTGCAGCAGCTGCCAGATGACCCCATTGGATCTATCGATGCACGATCTATCGAGT-GACGCTACCCA---GACTCCTCAGACTCCACAGACCGAAGACGCTCAGCCAGTGCAGCCCATTGAGGTGAGCATCGAACA-------CCATATATACTCGTAGTAGATTACCAATTAGCTACTTCAATCCGTGCCGCGCCCTGCCATAGAAATGCGAGCCTGAGCCGACCTCTAGCCAGGGCG----GGTCAGGTGAGCATCCTCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:40 PM