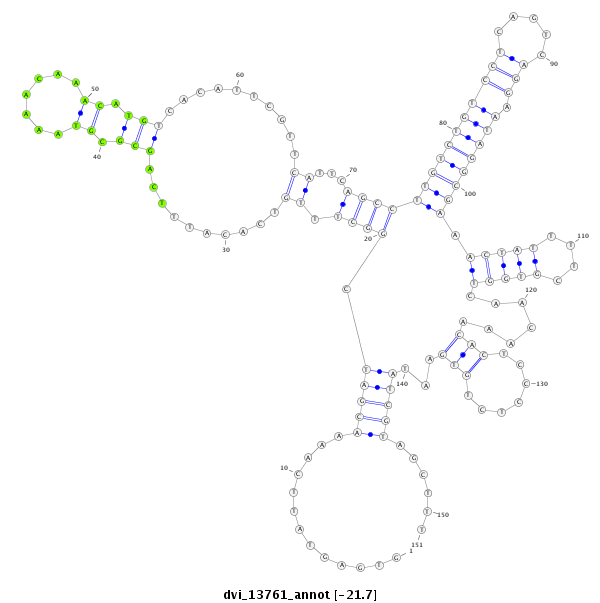
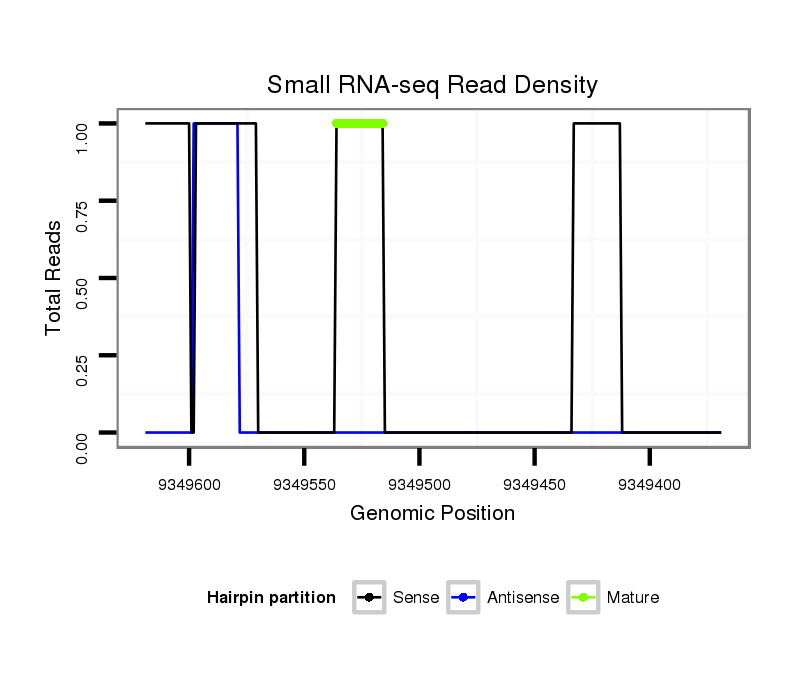
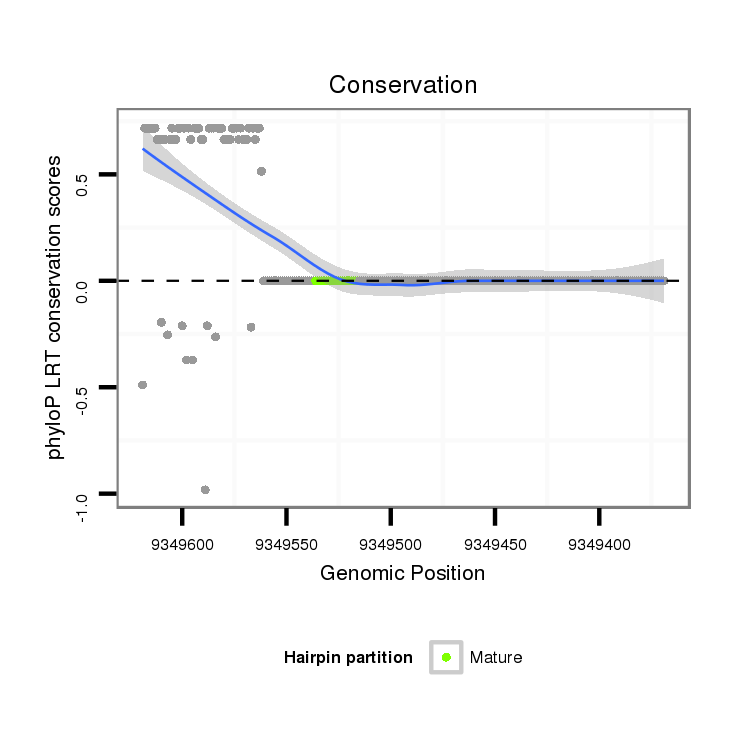
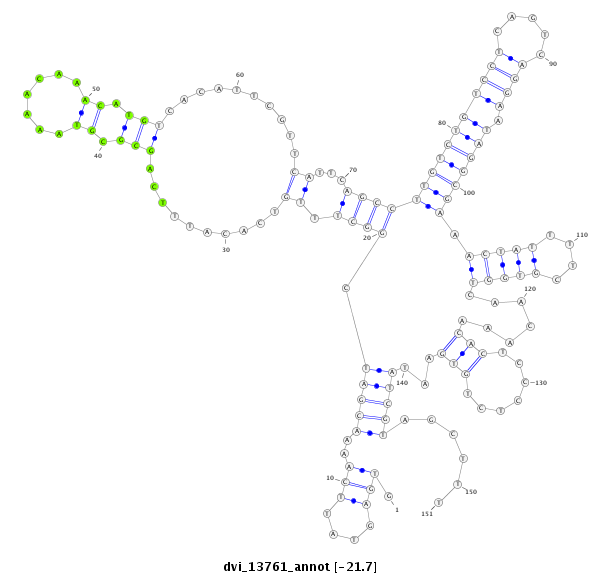
ID:dvi_13761 |
Coordinate:scaffold_12970:9349419-9349569 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.7 | -21.7 | -21.7 |
 |
 |
 |
exon [dvir_GLEANR_3521:3]; CDS [Dvir\GJ18697-cds]; intron [Dvir\GJ18697-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAATATCGCCGTGTGGATAACACGTTTCCCAAATTTATGCCGAATGACGGTGAGTATTCAAAACGATCGGCTTTGTCACATTTCAGCGCGTAAAACAAACATGTCACATTCGTTCATTCAGCCTTGTCTGTCCTCAGTCAGGAATAGGCGAAACTATTTTCGTGGTCAACAAACACTCCCTCTGTGAATATCGTAGCTTTGGGAGCTTGGCTATAGTAATTTGAGCTTCCTCCCCTCATCTTCGTTGTCC **************************************************.............(((((.((((.((..........(((.((.......)).)))..........))...))))(((((((((((.....))).)))))))).(((((....))))).......(((.......)))...)))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060655 9x160_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060683 160_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060659 Argentina_testes_total |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................GAATATCGTAGCTTTGGGAGC............................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| AAAATATCGCCGTGTGGATA....................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................ATTTACCCCGAATGACGGCGA..................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TCAGCGCGTAAAACAAACATG................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TATTTTGGAGGTCAACAAACA........................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................ACGTTTCCCAAATTTATGCCGAATGAC.......................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTCGTTGTCGACAAACCCTCC....................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TCGCCGTGTGGTTAACCGGT................................................................................................................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TCGCAGTGTGGATACCTCGTT................................................................................................................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................GCGTAAAACAGACATGTC................................................................................................................................................. | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................ACAGCAAACACTCCATCTGT................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................TATTTTGGAGGTCAACAAA............................................................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................AGTGATTAGAGCTTCGTCC................. | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................ATACCGTAACTTCGGGAG............................................. | 18 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTTTATAGCGGCACACCTATTGTGCAAAGGGTTTAAATACGGCTTACTGCCACTCATAAGTTTTGCTAGCCGAAACAGTGTAAAGTCGCGCATTTTGTTTGTACAGTGTAAGCAAGTAAGTCGGAACAGACAGGAGTCAGTCCTTATCCGCTTTGATAAAAGCACCAGTTGTTTGTGAGGGAGACACTTATAGCATCGAAACCCTCGAACCGATATCATTAAACTCGAAGGAGGGGAGTAGAAGCAACAGG
**************************************************.............(((((.((((.((..........(((.((.......)).)))..........))...))))(((((((((((.....))).)))))))).(((((....))))).......(((.......)))...)))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060689 160x9_testes_total |
M047 female body |
SRR060679 140x9_testes_total |
M061 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060658 140_ovaries_total |
V047 embryo |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................GTGCAAAGGGTTTAAATACG.................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................AACCGATATCATTCAACG.......................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................CAAAGGGTTTCAATAGGG................................................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CCAGGTGTTCGTGCGGGAGA................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GGAATAGACAAGAGTCAGTT............................................................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................AGTTGTTCGTGCGGGAGACG................................................................. | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................AGCAAGGAAGTCGGAGCAA.......................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................CAAAGGGTTTCAATAGGGG................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:9349369-9349619 - | dvi_13761 | AAAATATCGCCGTGTGGATAACACGTTTCCCAAATTTATGCCGAATGACGGTGAGTATTCAAAACGATCGGCTTTGTCACATTTCAGCGCGTAAAACAAACATGTCACATTCGTTCATTCAGCCTTGTCTGTCCTCAGTCAGGAATAGGCGAAACTATTTTCGTGGTCAACAAACACTCCCTCTGTGAATATCGTAGCTTTGGGAGCTTGGCTATAGTAATTTGAGCTTCCTCCCCTCATCTTCGTTGTCC |
| droMoj3 | scaffold_6308:1384002-1384058 + | CAAATATCGCCGCGTGGATAATACATTTCCTAAATATATGCCGAATGACGGTAAGTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15203:2036882-2036939 + | CAAATATCGTCGTGTGGATGATACATTTCCAGAATTTATGCCGAATGACGGTGAGTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 09:29 PM