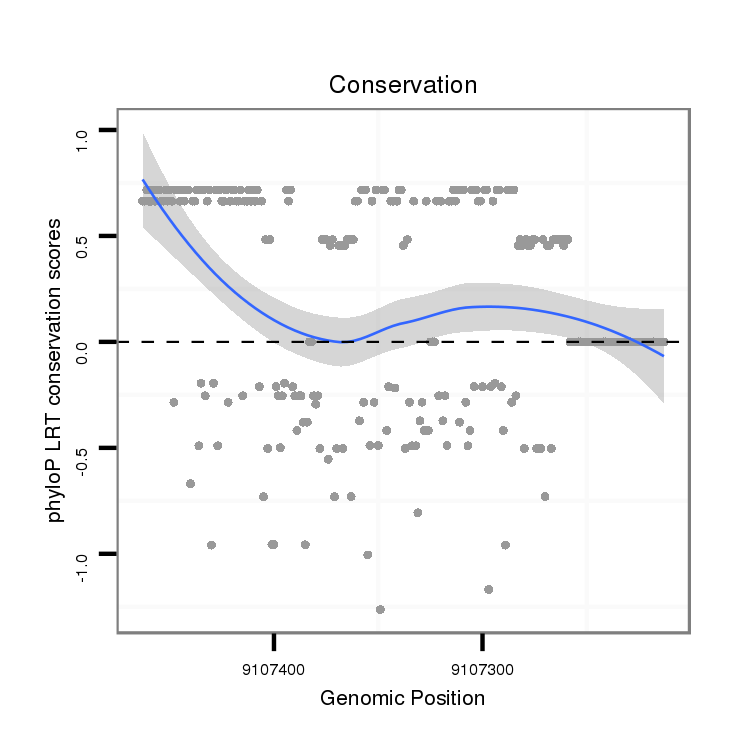
ID:dvi_13728 |
Coordinate:scaffold_12970:9107263-9107413 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ18715-cds]; exon [dvir_GLEANR_3541:1]; intron [Dvir\GJ18715-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCTGCAGTTGCTGTGTTACACATGCCTTCTACTTCTTTCGAAGATGCTAGGTGAGTACCTTTCCAAATGAGAATTTATTTAGTCTTTTTACATGGCCGAAGACGCTAATAGTTGCAAATGGGCAAGTTTTGTCCTTGAGCCGTCGACCGTGTAAATTTTATGCAAAATGGAAAAAATAAACACTTGCATGGTTCGGTAATAACAACCACAGTGCACTTATTGGGATCGACCGACTTAGAGATACCCTGAAC **************************************************.....((((...(((..((((..(((((((..(((.(((((((((.(((....(((.............((((((...))))))...)))..))).))))))))).............)))..)))))))..))))..)))...))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M028 head |
M047 female body |
SRR060687 9_0-2h_embryos_total |
SRR060673 9_ovaries_total |
V047 embryo |
SRR060681 Argx9_testes_total |
SRR060666 160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
M027 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................CAGTGCCGTTATTGGGAT......................... | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........GCTATGTTACACATTCCTTCGA............................................................................................................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................GAAGACGCTAATGATTGTAAAT.................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCCTTGAGAAGCCGACCGTG.................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................ACCTACTAGTTCTTTCGAAG................................................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TAACAGTCACAGTGCGCTTA................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................CTACTCCTTTGGAAGATCC............................................................................................................................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................GGTTGCAAATGGGAAAGT............................................................................................................................ | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................GTCCTTGTGGCGTCGACG....................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CGACGTCAACGACACAATGTGTACGGAAGATGAAGAAAGCTTCTACGATCCACTCATGGAAAGGTTTACTCTTAAATAAATCAGAAAAATGTACCGGCTTCTGCGATTATCAACGTTTACCCGTTCAAAACAGGAACTCGGCAGCTGGCACATTTAAAATACGTTTTACCTTTTTTATTTGTGAACGTACCAAGCCATTATTGTTGGTGTCACGTGAATAACCCTAGCTGGCTGAATCTCTATGGGACTTG
**************************************************.....((((...(((..((((..(((((((..(((.(((((((((.(((....(((.............((((((...))))))...)))..))).))))))))).............)))..)))))))..))))..)))...))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1106729 mixed whole adult body |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
GSM1528803 follicle cells |
V047 embryo |
M028 head |
M047 female body |
SRR060655 9x160_testes_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................AGGTACTCGGCAGGTGGC...................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TCAAAGCAGGAACTC................................................................................................................ | 15 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TGGTGGCACGTGAATAAG............................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................ACGCTAGCAGCCTGAATCT............ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CGGCAGCTGGCAGAGATAA.............................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGGTACCAAGCCATCATA................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CGGTGGTGGCACGTGAATA............................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................GTTGGCGTCACATGAGTAA.............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................GAAGTTGAAGAAGGTTTCTA.............................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12970:9107213-9107463 - | dvi_13728 | GCTGCAGTTGCTGTGTTACACATGCCTTCTACTTCTTTCGAAGATGCTAGGTGAGTACCTTTCCAAATGAGAATTTATTTAGTCTTTTTACATGGCCGAAGA---------CGCTAATAGTTGCAA------------------ATGGGCAAGTTTTGTCCTTGAGCCGTCGACCGTGTAAATTTTATGCAAAATGGAAAAAATAAACA--------------CTTGCATGGTTCGGTAATAACAACCACAGTGCACTTATTGGGATCGACCGACTTAGAGATACCCTGAAC |
| droMoj3 | scaffold_6473:16335750-16335966 + | GCTGCAGTTGCTGTGCTACACATTCCTGCTGCTACTGTCGAGGATGCTGGGTGAGTACATCTGGAGCCGAGAACACGTCT--CCCCTTTTCAGAGCTGAATA------ATTCGTTGAGCGCTGGAA------------------TTGGACAAGCTCGGGGTCAGT---GCCAGCCGTGTAAACGATATGCAAAGTGGAAATTATGAGCACCAGCTGACGAGCGCCTGCATAACTAGGCAATAACAA---------------------------------------------- | |
| droGri2 | scaffold_15203:11787654-11787829 - | GCTGCAGTTGCTGTGCTACACATTCCTGTTACTCTTGTCGAGGATGCTAGGTGAGTGC------GAATAAGAGTATAGAG--TAT-----------------AGCTTTGATCGTTGAACGCTCAAACGCCTGTGCACTGGGCCCTCGGGCAA---CGGGGTCAGT---GTCAAGCGTGTCAACGATGTGCGAACCGAAAGTGATGAA------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 09:08 PM